8V98
 
 | | GII.24 Loreto 1972 norovirus protruding domain | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Capsid protein VP1 | | Authors: | Kher, G, Prewitt, A, Pancera, M, Hansman, G. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of a broad-spectrum therapeutic Fc-nanobody for human noroviruses.
J.Virol., 98, 2024
|
|
8V46
 
 | | CryoEM structure of AriA-AriB complex (Form I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AriA antitoxin, AriB, ... | | Authors: | Deep, A, Corbett, K.D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Architecture and activation mechanism of the bacterial PARIS defence system.
Nature, 2024
|
|
8W0X
 
 | |
3MDK
 
 | |
3MDW
 
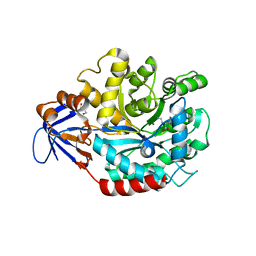 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | GLYCEROL, N-[(E)-iminomethyl]-L-aspartic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8979 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
8W0Y
 
 | |
8W8F
 
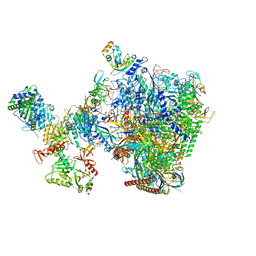 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
3MMR
 
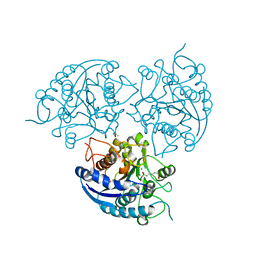 | | Structure of Plasmodium falciparum Arginase in complex with ABH | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase, BETA-MERCAPTOETHANOL, ... | | Authors: | Dowling, D.P, Ilies, M, Olszewski, K.L, Portugal, S, Mota, M.M, Llinas, M, Christianson, D.W. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of arginase from Plasmodium falciparum and implications for L-arginine depletion in malarial infection .
Biochemistry, 49, 2010
|
|
8W0W
 
 | |
3MOS
 
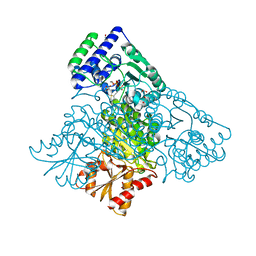 | | The structure of human Transketolase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Parthier, C, Tittmann, K. | | Deposit date: | 2010-04-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of human transketolase and new insights into its mode of action.
J.Biol.Chem., 285, 2010
|
|
8W0V
 
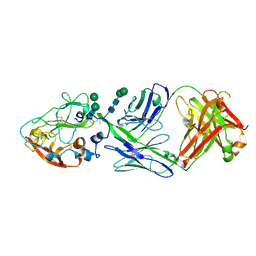 | |
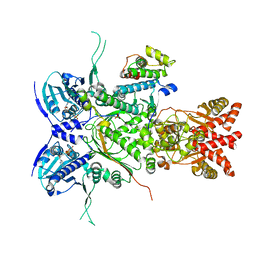
8U1L
 
 | |
3MI9
 
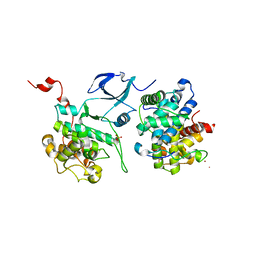 | | Crystal structure of HIV-1 Tat complexed with human P-TEFb | | Descriptor: | Cell division protein kinase 9, Cyclin-T1, Protein Tat, ... | | Authors: | Tahirov, T.H, Babayeva, N.D, Varzavand, K, Cooper, J.J, Sedore, S.C, Price, D.H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb.
Nature, 465, 2010
|
|
3MJL
 
 | |
8V8E
 
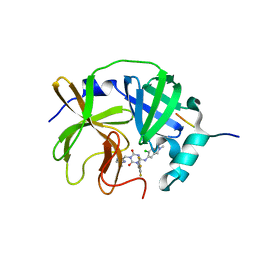 | | Room-temperature X-ray structure of SARS-CoV-2 main protease catalytic domain (residues 1-199-6H) in complex with ensitrelvir (ESV) | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, peptide | | Authors: | Kovalevsky, A, Coates, L. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Visualizing the Active Site Oxyanion Loop Transition Upon Ensitrelvir Binding and Transient Dimerization of SARS-CoV-2 Main Protease.
J.Mol.Biol., 436, 2024
|
|
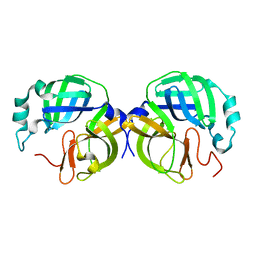
8V7W
 
 | |
9GPB
 
 | |
2VV6
 
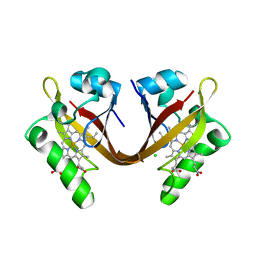 | | BJFIXLH IN FERRIC FORM | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL, ... | | Authors: | Ayers, R.A, Moffat, K. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Quaternary Structure in the Signaling Mechanisms of Pas Domains.
Biochemistry, 47, 2008
|
|
3M7F
 
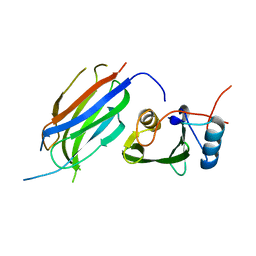 | | Crystal structure of the Nedd4 C2/Grb10 SH2 complex | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Growth factor receptor-bound protein 10 | | Authors: | Huang, Q, Szebenyi, M. | | Deposit date: | 2010-03-16 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Interaction between the Growth Factor-binding Protein GRB10 and the E3 Ubiquitin Ligase NEDD4.
J.Biol.Chem., 285, 2010
|
|
3ME3
 
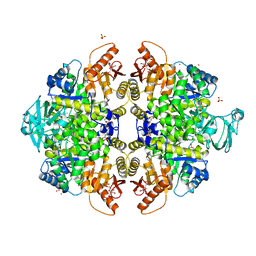 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-{[4-(2,3-dihydro-1,4-benzodioxin-6-ylsulfonyl)-1,4-diazepan-1-yl]sulfonyl}aniline, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
5A76
 
 | |
2VV8
 
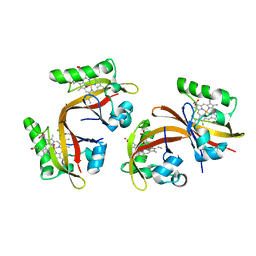 | | Co-bound structure of bjFixLH | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ayers, R.A, Moffat, K. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Changes in Quaternary Structure in the Signaling Mechanisms of Pas Domains.
Biochemistry, 47, 2008
|
|
3MDU
 
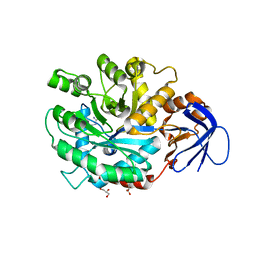 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutamate | | Descriptor: | GLYCEROL, N-carbamimidoyl-L-glutamic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4003 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
3MIA
 
 | | Crystal structure of HIV-1 Tat complexed with ATP-bound human P-TEFb | | Descriptor: | Cell division protein kinase 9, Cyclin-T1, MAGNESIUM ION, ... | | Authors: | Tahirov, T.H, Babayeva, N.D, Varzavand, K, Cooper, J.J, Sedore, S.C, Price, D.H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb.
Nature, 465, 2010
|
|
3MKH
 
 | | Podospora anserina Nitroalkane Oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, NITROALKANE OXIDASE, ... | | Authors: | Tormos, J.R, Taylor, A.B, Daubner, S.C, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2010-04-14 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Identification of a hypothetical protein from Podospora anserina as a nitroalkane oxidase.
Biochemistry, 49, 2010
|
|
