8TBU
 
 | | Structure of human erythrocyte pyruvate kinase in complex with an allosteric activator Compound 12 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-[(4-hydroxyphenyl)methyl]-2,4-dimethyl-4,6-dihydro-5H-[1,3]thiazolo[5',4':4,5]pyrrolo[2,3-d]pyridazin-5-one, MANGANESE (II) ION, ... | | Authors: | Jin, L, Padyana, A. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Design of AG-946, a Pyruvate Kinase Activator.
Chemmedchem, 19, 2024
|
|
6Y8Y
 
 | | Structure of Baltic Herring (Clupea Harengus) Phosphoglucomutase 5 (PGM5) with bound Glucose-1-Phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Gustafsson, R, Eckhard, U, Selmer, M. | | Deposit date: | 2020-03-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring-An Inactive Enzyme with Intact Substrate Binding.
Biomolecules, 10, 2020
|
|
7OL2
 
 | | Crystal structure of mouse contactin 1 immunoglobulin domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-1, ... | | Authors: | Chataigner, L.M.P, Janssen, B.J.C. | | Deposit date: | 2021-05-19 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structural insights into the contactin 1 - neurofascin 155 adhesion complex.
Nat Commun, 13, 2022
|
|
9BGG
 
 | | Structure of a hyperactive S1S3 truncation of the human GlcNAc-1-phosphotransferase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, highly active truncation of GlcNAc-1-phosphotransferase, S1S3,N-acetylglucosamine-1-phosphotransferase subunit beta | | Authors: | Li, H, Li, H. | | Deposit date: | 2024-04-18 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a truncated human GlcNAc-1-phosphotransferase variant reveals the basis for its hyperactivity.
J.Biol.Chem., 300, 2024
|
|
5MYV
 
 | | Crystal structure of SRPK2 in complex with compound 1 | | Descriptor: | 5-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]furan-2-carboxamide, DIMETHYL SULFOXIDE, SRSF protein kinase 2,SRSF protein kinase 2, ... | | Authors: | Chaikuad, A, Pike, A.C.W, Savitsky, P, von Delft, F, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-29 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease.
ACS Chem. Biol., 12, 2017
|
|
6QB4
 
 | | Mcl1-scFv complex with an indole acid inhibitor | | Descriptor: | 3-[3-[[(1~{R})-1,2,3,4-tetrahydronaphthalen-1-yl]oxy]propyl]-7-(1,3,5-trimethylpyrazol-4-yl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, scFv55 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8HAO
 
 | | Human parathyroid hormone receptor-1 dimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha, ... | | Authors: | Zhao, L, Xu, H.E, Yuan, Q. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1.
Acta Pharmacol.Sin., 44, 2023
|
|
5KB3
 
 | | 1.4 A resolution structure of Helicobacter Pylori MTAN in complexed with p-ClPh-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-chlorophenyl)sulfanyl]methyl}pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase, MAGNESIUM ION | | Authors: | Banco, M.T, Ronning, D.R. | | Deposit date: | 2016-06-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Neutron structures of the Helicobacter pylori 5'-methylthioadenosine nucleosidase highlight proton sharing and protonation states.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8QG7
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl-pent-4-ynyl-amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QG9
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl-propan-2-yl-amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
6AN3
 
 | | Crystal structure of H172A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant soaked with peptide (no CuH bound, no peptide bound) | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
8QGH
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl]amino]propyl]-3-[(azanylidene-$l^{4}-azanylidene)amino]propanamide | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGK
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl]amino]propyl]ethanamide | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
5MVV
 
 | | Crystal structure of Plasmodium falciparum actin I- gelsolin segment 1 -CdATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-1, CADMIUM ION, ... | | Authors: | Panneerselvam, S, Kumpula, E.-P, Kursula, I, Burkhardt, A, Meents, A. | | Deposit date: | 2017-01-17 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid cadmium SAD phasing at the standard wavelength (1 angstrom ).
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6QGD
 
 | | Structure of human Mcl-1 in complex with thienopyrimidine inhibitor | | Descriptor: | 2-[(6-ethyl-5-phenyl-thieno[2,3-d]pyrimidin-4-yl)amino]-3-oxidanyl-propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6T2K
 
 | | Furano[2,3-d]prymidine amides as Notum inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-(6-chloranyl-7-cyclopropyl-thieno[3,2-d]pyrimidin-4-yl)sulfanylethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Scaffold-hopping identifies furano[2,3-d]pyrimidine amides as potent Notum inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6YJR
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
9BV5
 
 | | Crystal structure of human CYP3A4 in complex with SJ000362065 | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 3A4, N-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]-2,2-dimethylpropanamide, ... | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
5MXD
 
 | | BACE-1 IN COMPLEX WITH LIGAND 32397778 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, ~{N},~{N}-dimethyl-2-pyrrolidin-1-yl-quinazolin-4-amine | | Authors: | Alexander, R. | | Deposit date: | 2017-01-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Human Beta Secretase 1 In Complex With Ligand 32397778
to be published
|
|
9BM6
 
 | |
9BMD
 
 | |
9BMM
 
 | |
7ALR
 
 | | Crystal structure of TD1-gatorbulin1 complex | | Descriptor: | (2~{R})-2-oxidanyl-2-[(6~{S},9~{S},12~{S},15~{S},17~{S})-6,10,12,17-tetramethyl-3-methylidene-7-oxidanyl-2,5,8,11,14-pentakis(oxidanylidene)-13-oxa-1,4,7,10-tetrazabicyclo[13.3.0]octadecan-9-yl]ethanamide, Designed Ankyrin Repeat Protein (DARPIN) D1, GLYCEROL, ... | | Authors: | Oliva, M.A, Diaz, J.F. | | Deposit date: | 2020-10-07 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Gatorbulin-1, a distinct cyclodepsipeptide chemotype, targets a seventh tubulin pharmacological site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
9BMH
 
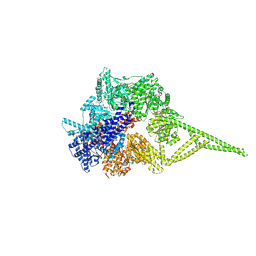 | |
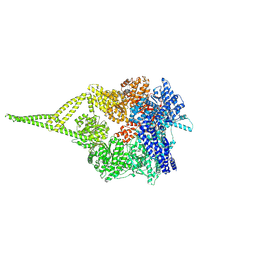
9BMY
 
 | |
