6DC4
 
 | | RSV-neutralizing human antibody AM22 | | Descriptor: | 1,2-ETHANEDIOL, Fab AM22 Heavy Chain, Fab AM22 Light Chain | | Authors: | Jones, H.G, McLellan, J.S. | | Deposit date: | 2018-05-04 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alternative conformations of a major antigenic site on RSV F.
Plos Pathog., 15, 2019
|
|
5B32
 
 | | The crystal structure of the heterotypic H2AZ/H2A nucleosome with H3.3. | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Horikoshi, N, Taguchi, H, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2016-02-08 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of heterotypic nucleosomes containing histones H2A.Z and H2A.
Open Biology, 6, 2016
|
|
6DA5
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-{[(2S)-1-oxo-3-phenyl-1-{[(pyridin-3-yl)methyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
6DAG
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-{[(2S)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}-3-phenylpropan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
6DC3
 
 | | RSV prefusion F bound to RSD5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab RSD5-Germline Heavy Chain, Fab RSD5-Germline Light Chain, ... | | Authors: | Battles, M.B, McLellan, J.S, Jones, H.J. | | Deposit date: | 2018-05-04 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Alternative conformations of a major antigenic site on RSV F.
Plos Pathog., 15, 2019
|
|
4KMH
 
 | |
4O6L
 
 | | Crystal Structure of TTK kinase domain with an inhibitor: 401498 (N-[(1R)-1-(2-chlorophenyl)propyl]-3-{4-[(1-methylpiperidin-4-yl)oxy]phenyl}-1H-indazole-5-carboxamide) | | Descriptor: | Dual specificity protein kinase TTK, GLYCEROL, N-[(1R)-1-(2-chlorophenyl)propyl]-3-{4-[(1-methylpiperidin-4-yl)oxy]phenyl}-1H-indazole-5-carboxamide | | Authors: | Qiu, W, Plotnikova, O, Feher, M, Awrey, D.E, Chirgadze, N.Y. | | Deposit date: | 2013-12-22 | | Release date: | 2014-12-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of TTK kinase domain with an inhibitor: 401498
TO BE PUBLISHED
|
|
6DRZ
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N-[(2S)-1-hydroxybutan-2-yl]-1,6-dimethyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WXN
 
 | | Structure of the LKB1 and 14-3-3 complex | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase STK11 | | Authors: | Ding, S, Shi, Z.B. | | Deposit date: | 2017-01-08 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure of the complex of phosphorylated liver kinase B1 and 14-3-3 zeta
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4OTN
 
 | |
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
6DRY
 
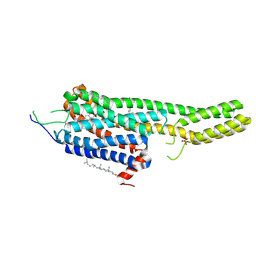 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5X0W
 
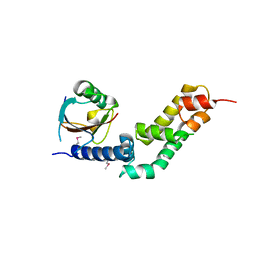 | | Molecular mechanism for the binding between Sharpin and HOIP | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Sharpin | | Authors: | Liu, J, Li, F, Cheng, X, Pan, L. | | Deposit date: | 2017-01-23 | | Release date: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into SHARPIN-Mediated Activation of HOIP for the Linear Ubiquitin Chain Assembly
Cell Rep, 21, 2017
|
|
5X1C
 
 | |
2Q80
 
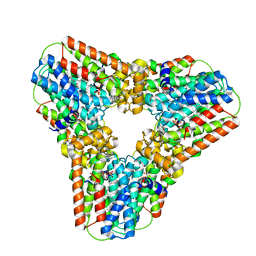 | | Crystal structure of human geranylgeranyl pyrophosphate synthase bound to GGPP | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Kavanagh, K.L, Dunford, J.E, Bunkoczi, G, Smee, C, von Delft, F, Arrowsmith, C, Weigelt, J, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-08 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of human geranylgeranyl pyrophosphate synthase reveals a novel hexameric arrangement and inhibitory product binding
J.Biol.Chem., 281, 2006
|
|
5X3P
 
 | |
5DX0
 
 | |
5X4L
 
 | | Crystal structure of the UBX domain of human UBXD7 in complex with p97 N domain | | Descriptor: | Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 7 | | Authors: | Jiang, T, Li, Z, Wang, Y, Xu, M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structures of the UBX domain of human UBXD7 and its complex with p97 ATPase
Biochem. Biophys. Res. Commun., 486, 2017
|
|
5DWQ
 
 | |
5XBF
 
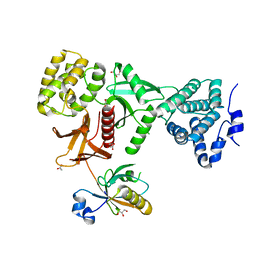 | | Crystal Structure of Myo7b C-terminal MyTH4-FERM in complex with USH1C PDZ3 | | Descriptor: | ACETATE ION, D-MALATE, GLYCEROL, ... | | Authors: | Li, J, He, Y, Weck, W.L, Lu, Q, Tyska, M.J, Zhang, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5X3S
 
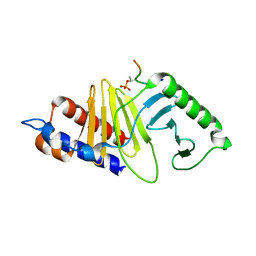 | |
6DXD
 
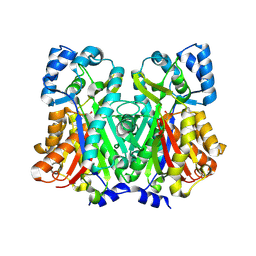 | |
6DO7
 
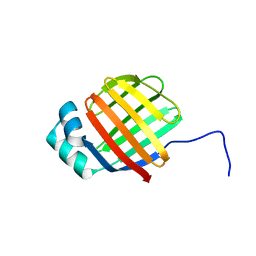 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
5E74
 
 | | Crystal Structure of BAZ2B bromodomain in complex with acetylindole compound UZH50 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, N-(1-acetyl-1H-indol-3-yl)-N-(5-hydroxy-2-methylphenyl)-3-(trifluoromethyl)benzamide | | Authors: | Lolli, G, Spiliotopoulos, D, Dolbois, A, Nevado, C, Caflisch, A. | | Deposit date: | 2015-10-11 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | The "Gatekeeper" Residue Influences the Mode of Binding of Acetyl Indoles to Bromodomains.
J. Med. Chem., 59, 2016
|
|
5X60
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 9 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
