7SV9
 
 | |
6QJQ
 
 | | Cryo-EM structure of heparin-induced 2N3R tau filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QKR
 
 | |
7TEB
 
 | |
7TA5
 
 | |
6QLG
 
 | | Crystal structure of AnUbiX (PadA1) in complex with FMN and dimethylallyl pyrophosphate | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYLALLYL DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Marshall, S.A, Payne, K.A.P, Leys, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The UbiX flavin prenyltransferase reaction mechanism resembles class I terpene cyclase chemistry.
Nat Commun, 10, 2019
|
|
7TE6
 
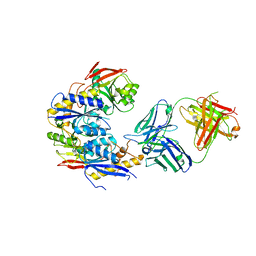 | | Crystal structure of GluN1b-2B ATD complexed to Fab5 anti-GluN2B antibody | | Descriptor: | Fab5 heavy chain, Fab5 light chain, Glutamate receptor ionotropic, ... | | Authors: | Regan, M, Tajima, N, Furukawa, H. | | Deposit date: | 2022-01-04 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.55 Å) | | Cite: | Development and characterization of functional antibodies targeting NMDA receptors.
Nat Commun, 13, 2022
|
|
6QLK
 
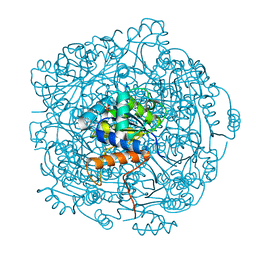 | | Crystal structure of F181H UbiX in complex with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Flavin prenyltransferase UbiX, PHOSPHATE ION, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The UbiX flavin prenyltransferase reaction mechanism resembles class I terpene cyclase chemistry.
Nat Commun, 10, 2019
|
|
6Q5Z
 
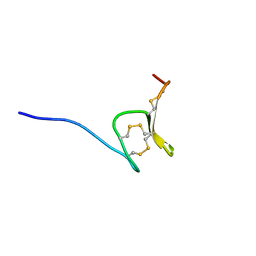 | | H-Vc7.2, H-superfamily conotoxin | | Descriptor: | Conotoxin Vc7.2 | | Authors: | Nielsen, L.D, Foged, M.M, Teilum, K, Ellgaard, L. | | Deposit date: | 2018-12-10 | | Release date: | 2019-04-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of an H-superfamily conotoxin reveals a granulin fold arising from a common ICK cysteine framework.
J.Biol.Chem., 294, 2019
|
|
7T00
 
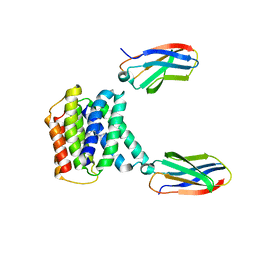 | |
7T5K
 
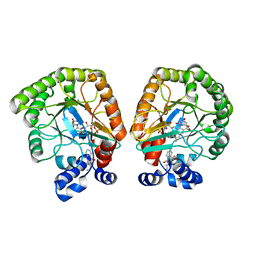 | | E. coli dihydroorotate dehydrogenase bound to the inhibitor HQNO | | Descriptor: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Horwitz, S.M, Ambarian, J.A, Davis, K.M. | | Deposit date: | 2021-12-12 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines.
Rsc Chem Biol, 3, 2022
|
|
6QJB
 
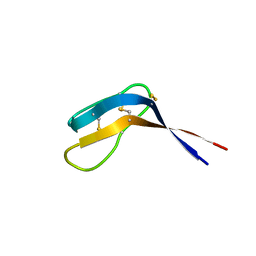 | | Truncated Evasin-3 (tEv3 17-56) | | Descriptor: | Evasin-3 | | Authors: | Denisov, S.S, Ippel, J.H, Heinzman, A.C.A, Koenen, R.R, Ortega-Gomez, A, Soehnlein, O, Hackeng, T.M, Dijkgraaf, I. | | Deposit date: | 2019-01-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Tick saliva protein Evasin-3 modulates chemotaxis by disrupting CXCL8 interactions with glycosaminoglycans and CXCR2.
J.Biol.Chem., 294, 2019
|
|
7TER
 
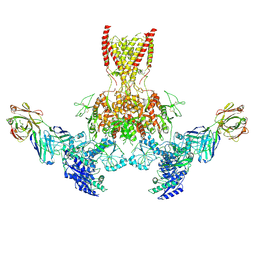 | |
6Q8M
 
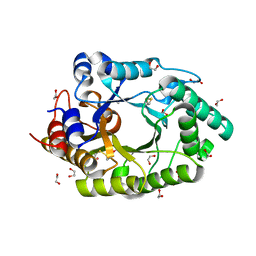 | | GH10 endo-xylanase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
7SU0
 
 | | Crystal structure of an acidic pH-selective Ipilimumab variant Ipi.105 in complex with CTLA-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, Cytotoxic T-lymphocyte protein 4, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Improved therapeutic index of an acidic pH-selective antibody.
Mabs, 14, 2022
|
|
6Q92
 
 | | Crystal structure of human Arginase-1 at pH 7.0 in complex with ABH | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase-1, MANGANESE (II) ION, ... | | Authors: | Grobben, Y, Uitdehaag, J.C.M, Zaman, G.J.R. | | Deposit date: | 2018-12-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into human Arginase-1 pH dependence and its inhibition by the small molecule inhibitor CB-1158.
J Struct Biol X, 4, 2020
|
|
7T3I
 
 | | CryoEM structure of the Rix7 D2 Walker B mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, Rix7, ... | | Authors: | Kocaman, S, Stanley, R.E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
6QJY
 
 | |
6QD4
 
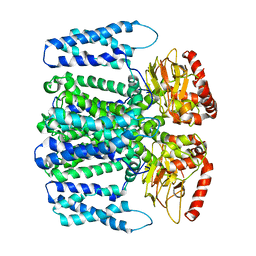 | | MloK1 model from single particle analysis of 2D crystals, class 8 (intermediate conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
7T6H
 
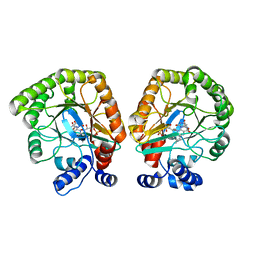 | | E. coli dihydroorotate dehydrogenase | | Descriptor: | Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, OROTIC ACID | | Authors: | Horwitz, S.M, Ambarian, J.A, Davis, K.M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines.
Rsc Chem Biol, 3, 2022
|
|
6Q97
 
 | |
7TBX
 
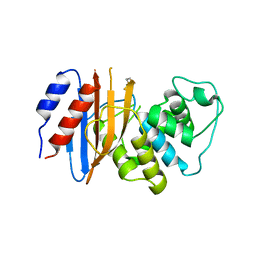 | | Crystal structure of D179Y KPC-2 beta-lactamase | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | van den Akker, F, Alsenani, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structural Characterization of the D179N and D179Y Variants of KPC-2 beta-Lactamase: Omega-Loop Destabilization as a Mechanism of Resistance to Ceftazidime-Avibactam.
Antimicrob.Agents Chemother., 66, 2022
|
|
7TEE
 
 | |
6QDK
 
 | |
6QE4
 
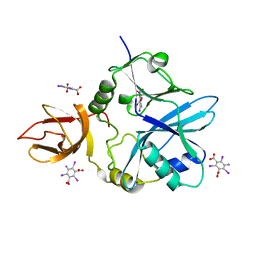 | | Re-refinement of 5OLI human IBA57-I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Putative transferase CAF17, mitochondrial | | Authors: | Calderone, V, Ciofi-Baffoni, S, Gourdoupis, S, Banci, L, Nasta, V. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In-house high-energy-remote SAD phasing using the magic triangle: how to tackle the P1 low symmetry using multiple orientations of the same crystal of human IBA57 to increase the multiplicity.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
