4RFL
 
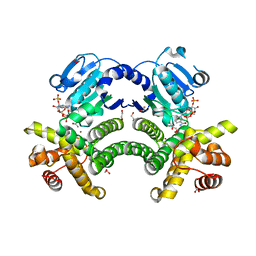 | | Crystal structure of G1PDH with NADPH from Methanocaldococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, Glycerol-1-phosphate dehydrogenase [NAD(P)+], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Carbone, V, Ronimus, R.S, Schofield, L.R, Sutherland-Smith, A.J. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Evolution of the Archaeal Lipid Synthesis Enzyme sn-Glycerol-1-phosphate Dehydrogenase.
J.Biol.Chem., 290, 2015
|
|
4CM3
 
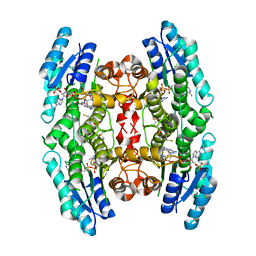 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 5-phenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
5TYR
 
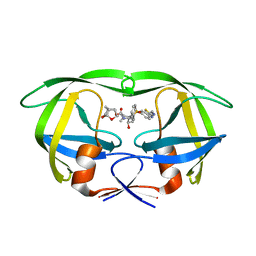 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
5TYS
 
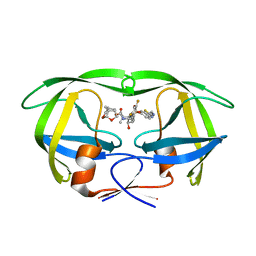 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
4PF4
 
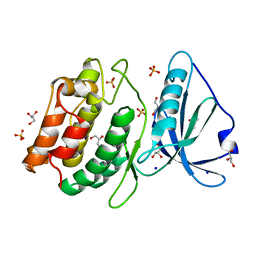 | | 1.1A X-RAY STRUCTURE OF THE APO CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE 1, aa 1-277 | | Descriptor: | Death-associated protein kinase 1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Temmerman, K, Simon, B, Wilmanns, M. | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | 1.1A X-RAY STRUCTURE OF THE APO CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE 1, aa 1-277
To Be Published
|
|
6A4Y
 
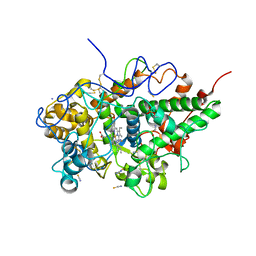 | | Crystal structure of bovine lactoperoxidase with partial occupancies of iodide and SCN- ions at the substrate binding site on the distal heme side at 1.92 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-06-21 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of bovine lactoperoxidase with partial occupancies of iodide and SCN- ions at the substrate binding site on the distal heme side at 1.92 A resolution
To Be Published
|
|
4LAE
 
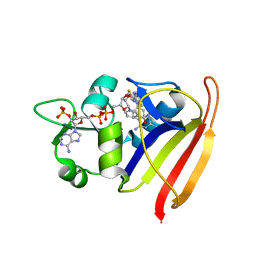 | | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines | | Descriptor: | 7-[5,6-dimethyl-2-(1,3-thiazol-2-yl)-1H-benzimidazol-1-yl]quinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M.T. | | Deposit date: | 2013-06-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines.
J.Med.Chem., 57, 2014
|
|
7XMG
 
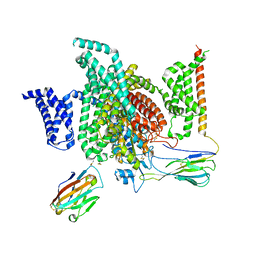 | | Cryo-EM structure of human NaV1.7/beta1/beta2-TCN-1752 | | Descriptor: | (1~{Z})-~{N}-[2-methyl-3-[(~{E})-[6-[4-[[4-(trifluoromethyloxy)phenyl]methoxy]piperidin-1-yl]-1~{H}-1,3,5-triazin-2-ylidene]amino]phenyl]ethanimidic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4JBE
 
 | | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, ... | | Authors: | Minasov, G, Filippova, E.V, Halavaty, A, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis.
TO BE PUBLISHED
|
|
2W0U
 
 | | CRYSTAL STRUCTURE OF HUMAN GLYCOLATE OXIDASE IN COMPLEX WITH THE INHIBITOR 5-[(4-CHLOROPHENYL)SULFANYL]- 1,2,3-THIADIAZOLE-4-CARBOXYLATE. | | Descriptor: | 5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole-4-carboxylate, FLAVIN MONONUCLEOTIDE, HYDROXYACID OXIDASE 1 | | Authors: | Bourhis, J.M, Lindqvist, Y. | | Deposit date: | 2008-10-10 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of Human Glycolate Oxidase in Complex with the Inhibitor 4-Carboxy-5-[(4-Chlorophenyl)Sulfanyl]-1,2,3-Thiadiazole.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6JO3
 
 | | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with substrate sn-glycerol-1-phosphate | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE | | Authors: | Nemoto, N, Miyazono, K, Tanokura, M, Yamagishi, A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with the substrate sn-glycerol 1-phosphate.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7FIQ
 
 | | The crystal structure of mannose-bound beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
6AYV
 
 | | Crystal structure of fructose-1,6-bisphosphatase T84A from Mycobacterium tuberculosis | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase class 2, GLYCEROL, ... | | Authors: | Abad-Zapatero, C, Wolf, N, Gutka, H.J, Movahedzadeh, F. | | Deposit date: | 2017-09-08 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Mycobacterium tuberculosis GlpX protein (class II fructose-1,6-bisphosphatase): implications for the active oligomeric state, catalytic mechanism and citrate inhibition.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5V36
 
 | | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-06 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD.
To Be Published
|
|
4LEK
 
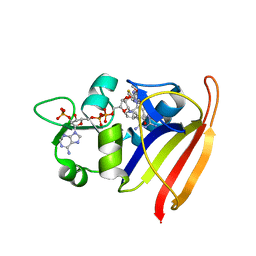 | | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines | | Descriptor: | 7-[5,6-dimethoxy-2-(1,3-thiazol-2-yl)-1H-benzimidazol-1-yl]quinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines.
J.Med.Chem., 57, 2014
|
|
6AYY
 
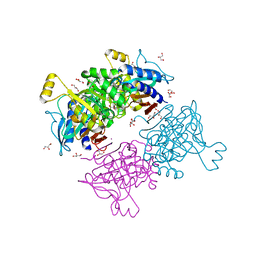 | | Crystal structure of Apo fructose-1,6-bisphosphatase from Mycobacterium tuberculosis | | Descriptor: | CITRIC ACID, Fructose-1,6-bisphosphatase class 2, GLYCEROL, ... | | Authors: | Abad-Zapatero, C, Wolf, N, Gutka, H.J, Movahedzadeh, F. | | Deposit date: | 2017-09-08 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structures of the Mycobacterium tuberculosis GlpX protein (class II fructose-1,6-bisphosphatase): implications for the active oligomeric state, catalytic mechanism and citrate inhibition.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6AYU
 
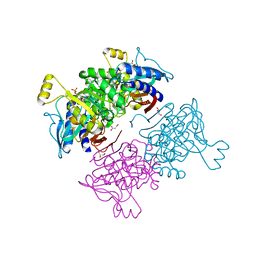 | | Crystal structure of fructose-1,6-bisphosphatase T84S from Mycobacterium tuberculosis | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase class 2, GLYCEROL, ... | | Authors: | Abad-Zapatero, C, Wolf, N, Gutka, H.J, Movahedzadeh, F. | | Deposit date: | 2017-09-08 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the Mycobacterium tuberculosis GlpX protein (class II fructose-1,6-bisphosphatase): implications for the active oligomeric state, catalytic mechanism and citrate inhibition.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4P38
 
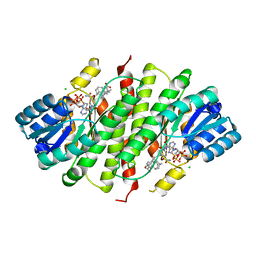 | | Human 11beta-Hydroxysteroid Dehydrogenase Type 1 in complex with AZD8329 | | Descriptor: | 4-[4-(2-adamantylcarbamoyl)-5-tert-butyl-pyrazol-1-yl]benzoic acid, CHLORIDE ION, Corticosteroid 11-beta-dehydrogenase isozyme 1, ... | | Authors: | Ogg, D, Hargreaves, D, Gerhardt, S. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel acidic 11 beta-hydroxysteroid dehydrogenase type 1 (11 beta-HSD1) inhibitor with reduced acyl glucuronide liability: the discovery of 4-[4-(2-adamantylcarbamoyl)-5-tert-butyl-pyrazol-1-yl]benzoic acid (AZD8329).
J.Med.Chem., 55, 2012
|
|
1M5W
 
 | | 1.96 A Crystal Structure of Pyridoxine 5'-Phosphate Synthase in Complex with 1-deoxy-D-xylulose phosphate | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, PHOSPHATE ION, Pyridoxal phosphate biosynthetic protein pdxJ | | Authors: | Yeh, J.I, Du, S, Pohl, E, Cane, D.E. | | Deposit date: | 2002-07-10 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multistate Binding in Pyridoxine 5'-Phosphate Synthase: 1.96 A Crystal Structure in
Complex with 1-deoxy-D-xylulose phosphate
Biochemistry, 41, 2002
|
|
5TIM
 
 | | REFINED 1.83 ANGSTROMS STRUCTURE OF TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE, CRYSTALLIZED IN THE PRESENCE OF 2.4 M-AMMONIUM SULPHATE. A COMPARISON WITH THE STRUCTURE OF THE TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE-GLYCEROL-3-PHOSPHATE COMPLEX | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Wierenga, R.K, Hol, W.G.J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex.
J.Mol.Biol., 220, 1991
|
|
2X2J
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha- 1,4-glucan lyase with deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
4OQM
 
 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with F-ARA-EdU | | Descriptor: | 1-(2-deoxy-2-fluoro-beta-D-arabinofuranosyl)-5-ethynylpyrimidine-2,4(1H,3H)-dione, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Neef, A.B, Westermaier, Y, Perozzo, R, Luedtke, N, Scapozza, L. | | Deposit date: | 2014-02-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with F-ARA-EdU
To be Published
|
|
4OOF
 
 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase W203F mutant bound to fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Allen, C.L, Kholodar, S.A, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alteration of the Flexible Loop in 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase Boosts Enthalpy-Driven Inhibition by Fosmidomycin.
Biochemistry, 53, 2014
|
|
4CLE
 
 | |
4RFB
 
 | | 1.93 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with Sialyl-Lewis X. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E.V, Halavaty, A, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 1.93 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with Sialyl-Lewis X.
TO BE PUBLISHED
|
|
