1NYU
 
 | |
2Q7Z
 
 | | Solution Structure of the 30 SCR domains of human Complement Receptor 1 | | Descriptor: | Complement receptor type 1 | | Authors: | Furtado, P.B, Huang, C.Y, Ihyembe, D, Hammond, R.A, Marsh, H.C, Perkins, S.J. | | Deposit date: | 2007-06-08 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | The Partly Folded Back Solution Structure Arrangement of the 30 SCR Domains in Human Complement Receptor Type 1 (CR1) Permits Access to its C3b and C4b Ligands
J.Mol.Biol., 375, 2008
|
|
3C8Y
 
 | | 1.39 Angstrom crystal structure of Fe-only hydrogenase | | Descriptor: | 2 IRON/2 SULFUR/3 CARBONYL/2 CYANIDE/WATER/METHYLETHER CLUSTER, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Pandey, A.S, Lemon, B.J, Peters, J.W. | | Deposit date: | 2008-02-14 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Dithiomethylether as a ligand in the hydrogenase h-cluster.
J.Am.Chem.Soc., 130, 2008
|
|
1NYS
 
 | |
3QZZ
 
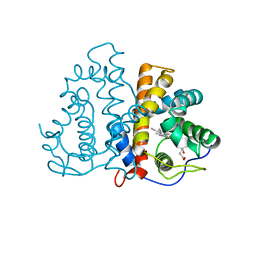 | | 3D Structure of Ferric Methanosarcina Acetivorans Protoglobin Y61W mutant in Aquomet form | | Descriptor: | Methanosarcina acetivorans protoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Tilleman, L, Dewilde, S, Ascenzi, P, Coletta, M, Ciaccio, C, Bruno, S, Moens, L, Bolognesi, M, Nardini, M. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural heterogeneity and ligand gating in ferric methanosarcina acetivorans protoglobin mutants.
Iubmb Life, 63, 2011
|
|
3KQR
 
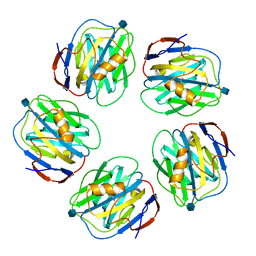 | | The structure of serum amyloid p component bound to phosphoethanolamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, ... | | Authors: | Mikolajek, H, Kolstoe, S.E, Wood, S.P, Pepys, M.B. | | Deposit date: | 2009-11-17 | | Release date: | 2010-12-08 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of ligand specificity in the human pentraxins, C-reactive protein and serum amyloid P component.
J.Mol.Recognit., 24, 2011
|
|
2YSA
 
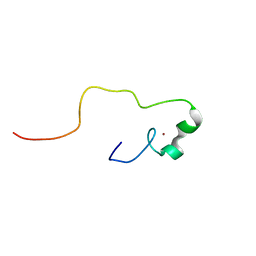 | | Solution structure of the zinc finger CCHC domain from the human retinoblastoma-binding protein 6 (Retinoblastoma-binding Q protein 1, RBQ-1) | | Descriptor: | Retinoblastoma-binding protein 6, ZINC ION | | Authors: | Ohnishi, S, Sato, M, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zinc finger CCHC domain from the human retinoblastoma-binding protein 6 (Retinoblastoma-binding Q protein 1, RBQ-1)
To be Published
|
|
4EH2
 
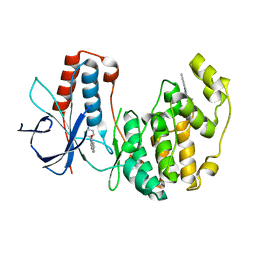 | | Human p38 MAP kinase in complex with NP-F1 and RL87 | | Descriptor: | 3-phenylquinazolin-4(3H)-one, Mitogen-activated protein kinase 14, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | Authors: | Over, B, Gruetter, C, Waldmann, H, Rauh, D. | | Deposit date: | 2012-04-02 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Natural-product-derived fragments for fragment-based ligand discovery.
Nat Chem, 5, 2012
|
|
4CDY
 
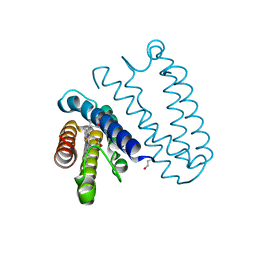 | | Spectroscopically-validated structure of cytochrome c prime from Alcaligenes xylosoxidans, reduced by X-ray irradiation at 160K | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-11-07 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8WW2
 
 | | GPR3/Gs complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Xiong, Y. | | Deposit date: | 2023-10-24 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Identification of oleic acid as an endogenous ligand of GPR3.
Cell Res., 34, 2024
|
|
1R56
 
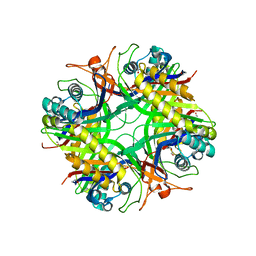 | | UNCOMPLEXED URATE OXIDASE FROM ASPERGILLUS FLAVUS | | Descriptor: | DI(HYDROXYETHYL)ETHER, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Prange, T. | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1Z9J
 
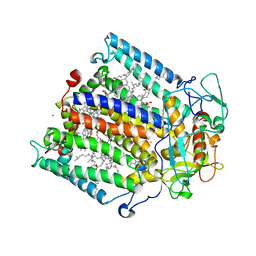 | | Photosynthetic Reaction Center from Rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Thielges, M, Uyeda, G, Camara-Artigas, A, Kalman, L, Williams, J.C, Allen, J.P. | | Deposit date: | 2005-04-02 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Design of a Redox-Linked Active Metal Site: Manganese Bound to Bacterial Reaction Centers at a Site Resembling That of Photosystem II
Biochemistry, 44, 2005
|
|
8WSS
 
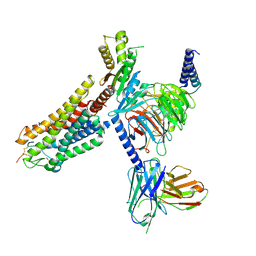 | | Cryo-EM structure of Melanin-Concentrating Hormone Receptor 1 with MCH | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, Shan, H, Hu, W, Wu, K, Xu, H.E, Zhang, Y, Wu, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Mechanisms of ligand recognition and activation of melanin-concentrating hormone receptors.
Cell Discov, 10, 2024
|
|
4CDV
 
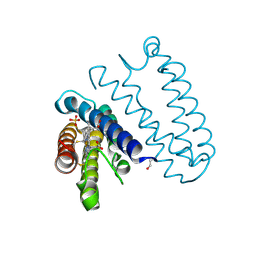 | | Spectroscopically-validated structure of cytochrome c prime from Alcaligenes xylosoxidans, reduced by X-ray irradiation at 100K | | Descriptor: | CYTOCHROME C', HEME C, SULFATE ION | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-11-06 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6B3W
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit,Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
3VQI
 
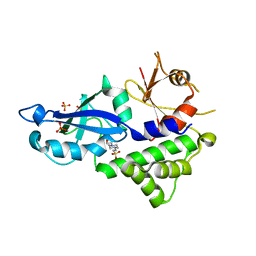 | | Crystal structure of Kluyveromyces marxianus Atg5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Atg5, SULFATE ION | | Authors: | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-03-24 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
4EH7
 
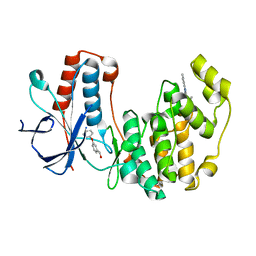 | | Human p38 MAP kinase in complex with NP-F6 and RL87 | | Descriptor: | (3-phenoxyphenyl)methanol, Mitogen-activated protein kinase 14, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | Authors: | Over, B, Gruetter, C, Waldmann, H, Rauh, D. | | Deposit date: | 2012-04-02 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natural-product-derived fragments for fragment-based ligand discovery.
Nat Chem, 5, 2012
|
|
3MXH
 
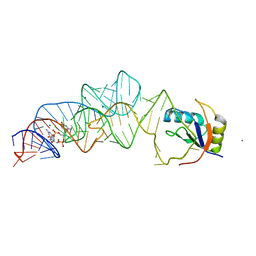 | | Native structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-07 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3R0G
 
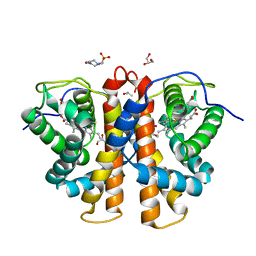 | | 3D Structure of Ferric Methanosarcina Acetivorans Protoglobin I149F mutant in Aquomet form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Pesce, A, Tilleman, L, Dewilde, S, Ascenzi, P, Coletta, M, Ciaccio, C, Bruno, S, Moens, L, Bolognesi, M, Nardini, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural heterogeneity and ligand gating in ferric methanosarcina acetivorans protoglobin mutants.
Iubmb Life, 63, 2011
|
|
4K71
 
 | | Crystal structure of a high affinity Human Serum Albumin variant bound to the Neonatal Fc Receptor | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, SULFATE ION, ... | | Authors: | Schmidt, M.M, Townson, S.A, Andreucci, A, Dombrowski, C, Erbe, D.V, King, B, Kovalchin, J.T, Masci, A, Murillo, A, Schirmer, E.B, Furfine, E.S, Barnes, T.M. | | Deposit date: | 2013-04-16 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an HSA/FcRn complex reveals recycling by competitive mimicry of HSA ligands at a pH-dependent hydrophobic interface.
Structure, 21, 2013
|
|
5XSW
 
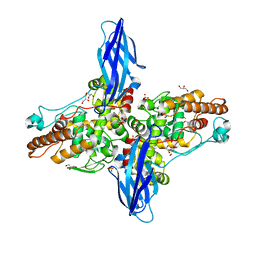 | |
3OEB
 
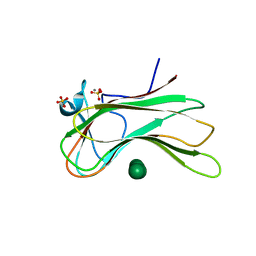 | |
4C7H
 
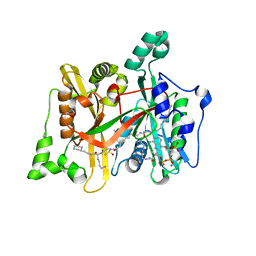 | | Leismania major N-myristoyltransferase in complex with a peptidomimetic (-NH2) molecule | | Descriptor: | COENZYME A, DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Olaleye, T.O, Brannigan, J.A, Goncalves, V, Roberts, S.M, Leatherbarrow, R.J, Wilkinson, A.J, Tate, E.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Peptidomimetic Inhibitors of N-Myristoyltransferase from Human Malaria and Leishmaniasis Parasites.
Org.Biomol.Chem., 12, 2014
|
|
5CF3
 
 | | Crystal structures of Bbp from Staphylococcus aureus | | Descriptor: | Bone sialoprotein-binding protein, CALCIUM ION | | Authors: | Yu, Y, Zhang, X.Y, Gu, J.K. | | Deposit date: | 2015-07-08 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Crystal structures of Bbp from Staphylococcus aureus reveal the ligand binding mechanism with Fibrinogen alpha
Protein Cell, 6, 2015
|
|
2YUR
 
 | | Solution structure of the Ring finger of human Retinoblastoma-binding protein 6 | | Descriptor: | Retinoblastoma-binding protein 6, ZINC ION | | Authors: | Abe, H, Miyamoto, K, Tochio, N, Tomizawa, T, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ring finger of human Retinoblastoma-binding protein 6
To be Published
|
|
