4U5T
 
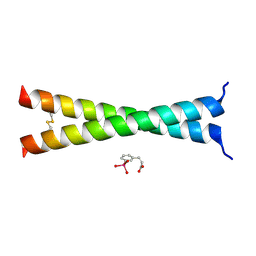 | | Crystal Structure of VBP Leucine Zipper with Bound Arylstibonic Acid | | Descriptor: | (2Z)-3-{3-[dihydroxy(oxido)-lambda~5~-stibanyl]phenyl}prop-2-enoic acid, VBP leucine zipper | | Authors: | Stagno, J.R, Ji, X. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | P6981, an arylstibonic acid, is a novel low nanomolar inhibitor of cAMP response element-binding protein binding to DNA.
Mol.Pharmacol., 82, 2012
|
|
6DPN
 
 | |
7R8J
 
 | |
7R8G
 
 | |
7R8H
 
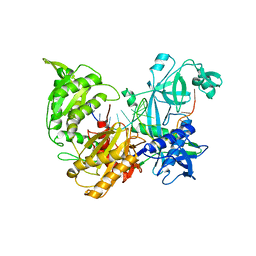 | |
8HEV
 
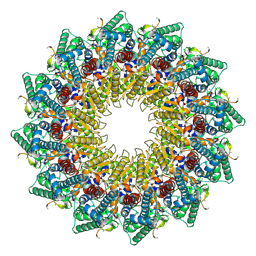 | | C12 portal in HCMV B-capsid | | Descriptor: | Portal protein, Unknown peptide | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HEU
 
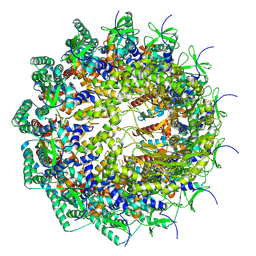 | | C12 portal in HCMV A-capsid | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
6DIE
 
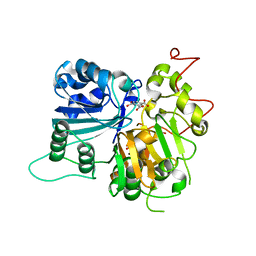 | | Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment benzene-1,2,4-tricarboxylic acid from single soak | | Descriptor: | 1,2-ETHANEDIOL, Tdp1 catalytic domain (residues 149-609), benzene-1,2,4-tricarboxylic acid | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-23 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
5W0U
 
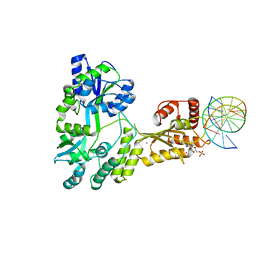 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5W1C
 
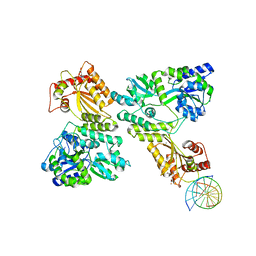 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
8H1J
 
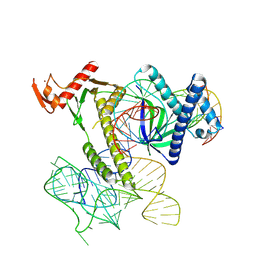 | | Cryo-EM structure of the TnpB-omegaRNA-target DNA ternary complex | | Descriptor: | Non-target strand, RNA-guided DNA endonuclease TnpB, Target strand, ... | | Authors: | Nakagawa, R, Hirano, H, Omura, S, Nureki, O. | | Deposit date: | 2022-10-03 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the transposon-associated TnpB enzyme.
Nature, 616, 2023
|
|
2GB9
 
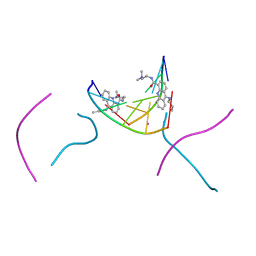 | | d(CGTACG)2 crosslinked bis-acridine complex | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9,9'-(HEXANE-1,6-DIYLDIIMINO)BIS{N-[2-(DIMETHYLAMINO)ETHYL]ACRIDINE-4-CARBOXAMIDE}, STRONTIUM ION | | Authors: | Hopcroft, N.H, Brogden, A.L, Searcey, M, Cardin, C.J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystallographic study of DNA duplex cross-linking: simultaneous binding to two d(CGTACG)2 molecules by a bis(9-aminoacridine-4-carboxamide) derivative.
Nucleic Acids Res., 34, 2006
|
|
6DLK
 
 | |
6DLY
 
 | |
8H9D
 
 | | Crystal structure of Cas12a protein | | Descriptor: | Cas12A, MAGNESIUM ION, RNA (5'-R(P*AP*AP*UP*UP*UP*CP*UP*AP*CP*UP*AP*AP*GP*UP*GP*UP*AP*GP*AP*UP*C)-3'), ... | | Authors: | Jianwei, L, Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of apo Cas12a and its complex with crRNA and DNA reveal the dynamics of ternary complex formation and target DNA cleavage.
Plos Biol., 21, 2023
|
|
5E5P
 
 | | Wild type I-SmaMI in the space group of C121 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, I-SmaMI LAGLIDADG meganuclease | | Authors: | Shen, B.W. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
1I4V
 
 | |
7RCF
 
 | |
7RCG
 
 | |
7RCD
 
 | |
7RCE
 
 | |
7RCC
 
 | |
8GIS
 
 | |
8GIR
 
 | |
8GIO
 
 | |
