7K03
 
 | |
7K1P
 
 | |
7K27
 
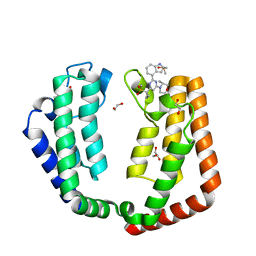 | | Crystal structure of the tandem bromodomain (BD1, BD2) of human TAF1 bound to ATR inhibitor AZ20 | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[(3R)-3-methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-yl}-1H-indole, DIMETHYL SULFOXIDE, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-09-08 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
7K3O
 
 | |
7K6F
 
 | | Crystal structure of the tandem bromodomain (BD1, BD2) of human TAF1 in complex with MES (2-(N-morpholino)ethanesulfonic acid) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2020-09-20 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
7K42
 
 | | Crystal structure of the second bromodomain (BD2) of human TAF1 bound to dioxane | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, SULFATE ION, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
5FYQ
 
 | | Sirt2 in complex with a 13-mer trifluoroacetylated Ran peptide | | Descriptor: | NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, RAN AA 31-43, SULFATE ION, ... | | Authors: | Knyphausen, P, de Boor, S, Scislowski, L, Extra, A, Baldus, L, Schacherl, M, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights Into Lysine-Deacetylation of Natively Folded Substrate Proteins by Sirtuins.
J.Biol.Chem., 291, 2016
|
|
3QWP
 
 | | Crystal structure of SET and MYND domain containing 3; Zinc finger MYND domain-containing protein 1 | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, SET and MYND domain-containing protein 3, ... | | Authors: | Dong, A, Dombrovski, L, Li, Y, Wernimont, A, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-28 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Crystal Structure of human Histone-lysine N-methyltransferase SMYD3
To be Published
|
|
5FLV
 
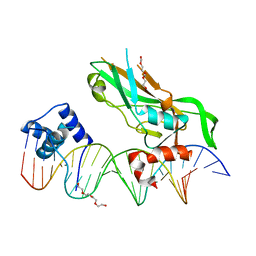 | | Crystal structure of NKX2-5 and TBX5 bound to the Nppa promoter region | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*TP*TP*CP*AP*AP*AP*GP*GP*TP *GP*TP*GP*AP*GP*AP*AP*G)-3', 5'-D(*TP*CP*TP*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP *TP*GP*AP*AP*GP*TP*GP*G)-3', HOMEOBOX PROTEIN NKX-2.5, ... | | Authors: | Stirnimann, C.U, Glatt, S, Mueller, C.W. | | Deposit date: | 2015-10-28 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis.
Cell(Cambridge,Mass.), 164, 2016
|
|
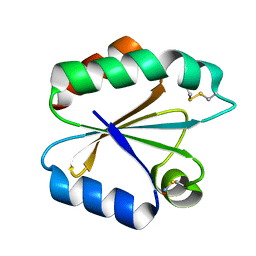
4OO5
 
 | |
8TXV
 
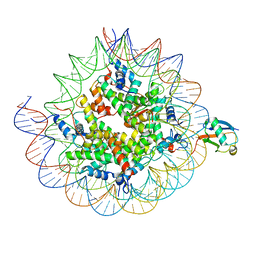 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXX
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 3) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXW
 
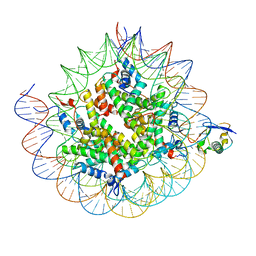 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
5EGA
 
 | | 2009 H1N1 PA endonuclease domain in complex with an N-acylhydrazone inhibitor | | Descriptor: | 3,4,5-tris(oxidanyl)-~{N}-[(~{E})-[3,4,5-tris(oxidanyl)phenyl]methylideneamino]benzamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-10-26 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | N-acylhydrazone inhibitors of influenza virus PA endonuclease with versatile metal binding modes.
Sci Rep, 6, 2016
|
|
8U8J
 
 | |
8U8K
 
 | |
7JJH
 
 | |
7JJG
 
 | |
7BDQ
 
 | | MAPK14 bound with SR300 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 14, ... | | Authors: | Schroeder, M, Roehm, S, Joerger, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
7BE4
 
 | | Crystal structure of MAP kinase p38 alpha in complex with inhibitor SR159 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{R})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
2Y9Q
 
 | | Crystal structure of human ERK2 complexed with a MAPK docking peptide | | Descriptor: | MAP KINASE-INTERACTING SERINE/THREONINE-PROTEIN KINASE 1, MITOGEN-ACTIVATED PROTEIN KINASE 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Barkai, T, Garai, A, Toeroe, I, Remenyi, A. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Specificity of Linear Motifs that Bind to a Common Mitogen-Activated Protein Kinase Docking Groove.
Sci. Signal, 5, 2012
|
|
7BE5
 
 | | Crystal structure of MAP kinase p38 alpha in complex with inhibitor SR276 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{R})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-methyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8000524 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
7BDO
 
 | | MAPK14 bound with SR302 | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{S})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Schroeder, M, Roehm, S, Joerger, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
8ODR
 
 | | Mimetic of UBC9-SUMO1 | | Descriptor: | SULFATE ION, SUMO-conjugating enzyme UBC9, Small ubiquitin-related modifier 1 | | Authors: | Coste, F, Goffinont, S, Suskiewicz, M.J. | | Deposit date: | 2023-03-09 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into the regulation of the human E2∼SUMO conjugate through analysis of its stable mimetic.
J.Biol.Chem., 299, 2023
|
|
8UPF
 
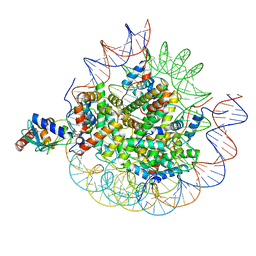 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168-UbcH5c | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
