7ZVT
 
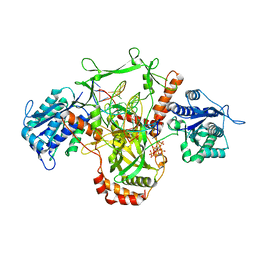 | | CryoEM structure of Ku heterodimer bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7ZWA
 
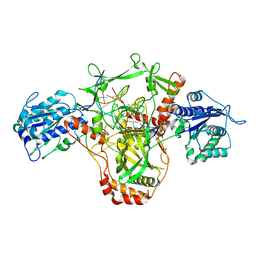 | | CryoEM structure of Ku heterodimer bound to DNA and PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
7ZYG
 
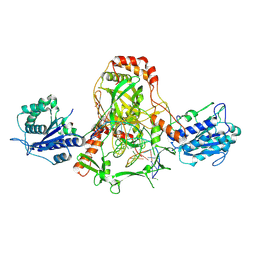 | | CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF | | Descriptor: | DNA, Non-homologous end-joining factor 1, PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
5E6A
 
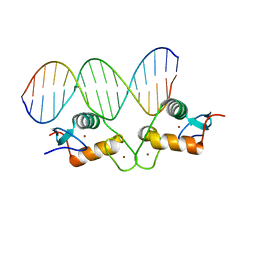 | | Glucocorticoid receptor DNA binding domain - PLAU NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*TP*GP*AP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
5E6B
 
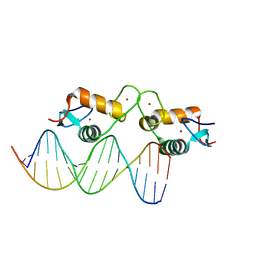 | | Glucocorticoid receptor DNA binding domain - RELB NF-kB response element complex | | Descriptor: | DNA (5'-D(*CP*CP*GP*GP*GP*GP*AP*AP*TP*TP*CP*CP*GP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*CP*GP*GP*AP*AP*TP*TP*CP*CP*CP*CP*GP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
5E6C
 
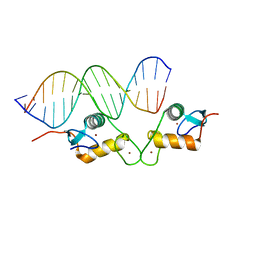 | | Glucocorticoid receptor DNA binding domain - CCL2 NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*GP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
5E69
 
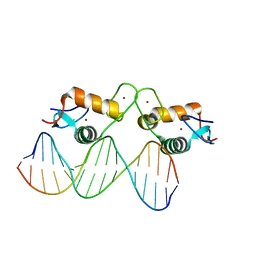 | | Glucocorticoid receptor DNA binding domain - IL8 NF-kB response element complex | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*CP*GP*AP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
5E6D
 
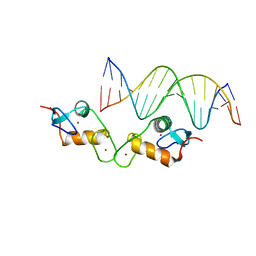 | | Glucocorticoid receptor DNA binding domain - ICAM1 NF-kB response element complex | | Descriptor: | DNA (5'-D(*GP*CP*TP*CP*CP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*GP*GP*AP*GP*C)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
4P9U
 
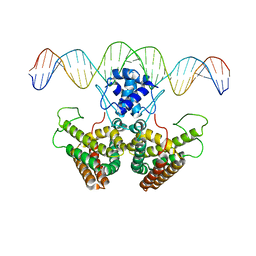 | |
3CFO
 
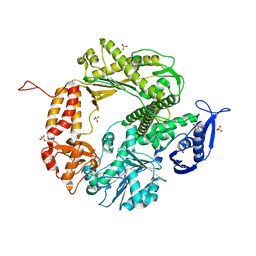 | | Triple Mutant APO structure | | Descriptor: | DNA polymerase, GUANOSINE, SULFATE ION | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
2R5Z
 
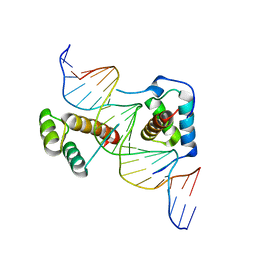 | | Structure of Scr/Exd complex bound to a DNA sequence derived from the fkh gene | | Descriptor: | DNA (5'-D(*DAP*DCP*DTP*DCP*DTP*DAP*DAP*DGP*DAP*DTP*DTP*DAP*DAP*DTP*DCP*DGP*DGP*DCP*DTP*DG)-3'), DNA (5'-D(*DTP*DCP*DAP*DGP*DCP*DCP*DGP*DAP*DTP*DTP*DAP*DAP*DTP*DCP*DTP*DTP*DAP*DGP*DAP*DG)-3'), Homeobox protein extradenticle, ... | | Authors: | Aggarwal, A.K, Passner, J.M, Jain, R. | | Deposit date: | 2007-09-04 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional specificity of a Hox protein mediated by the recognition of minor groove structure
Cell(Cambridge,Mass.), 131, 2007
|
|
7RVA
 
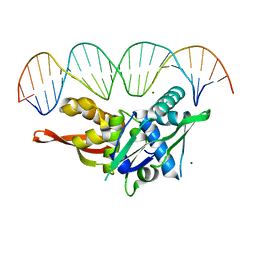 | | Updated Crystal Structure of Replication Initiator Protein REPE54. | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*T)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Stabilizing DNA-Protein Co-Crystals via Intra-Crystal Chemical Ligation of the DNA
Crystals, 12, 2022
|
|
7VO9
 
 | |
7S81
 
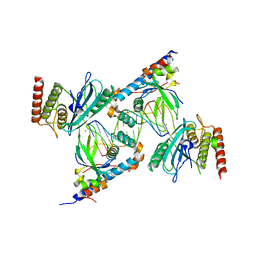 | | Structure of human PARP1 domains (Zn1, Zn3, WGR, HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*GP*GP*CP*CP*GP*CP*AP*T)-3'), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
2WB1
 
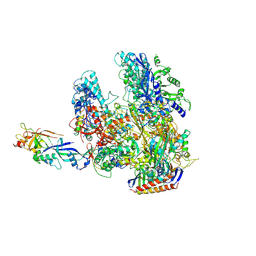 | | The complete structure of the archaeal 13-subunit DNA-directed RNA Polymerase | | Descriptor: | DNA-DIRECTED RNA POLYMERASE RPO10 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO11 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO12 SUBUNIT, ... | | Authors: | Korkhin, Y, Unligil, U.M, Littlefield, O, Nelson, P.J, Stuart, D.I, Sigler, P.B, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Evolution of Complex RNA Polymerase: The Complete Archaeal RNA Polymerase Structure
Plos Biol., 7, 2009
|
|
4PT4
 
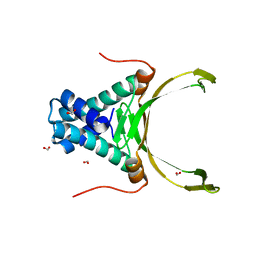 | | Crystal structure Analysis of N terminal region containing the dimerization domain and DNA binding domain of HU protein(Histone like protein-DNA binding) from Mycobacterium tuberculosis [H37Ra] | | Descriptor: | DNA-binding protein HU homolog, FORMIC ACID | | Authors: | Bhowmick, T, Ramagopal, U.A, Ghosh, S, Nagaraja, V, Ramakumar, S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Targeting Mycobacterium tuberculosis nucleoid-associated protein HU with structure-based inhibitors
Nat Commun, 5, 2014
|
|
8E9A
 
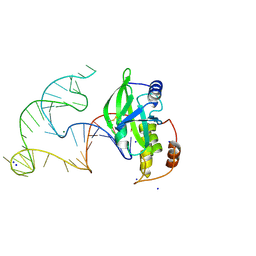 | | Crystal structure of AsfvPolX in complex with 10-23 DNAzyme and Mg | | Descriptor: | DNA/RNA (52-MER), MAGNESIUM ION, Repair DNA polymerase X, ... | | Authors: | Cramer, E.R, Robart, A.R, Kaya, A.I. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of a 10-23 deoxyribozyme exhibiting a homodimer conformation.
Commun Chem, 6, 2023
|
|
6SKO
 
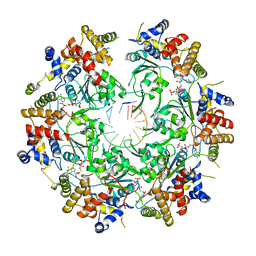 | | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork - conformation 2 MCM CTD:ssDNA | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
7KSX
 
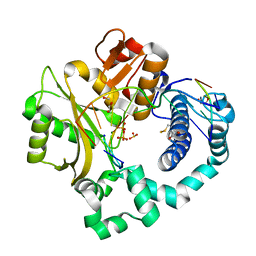 | | DNA Polymerase Mu, dGTP:Ct Product State Ternary Complex, 10 mM Mg2+ (30min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KSW
 
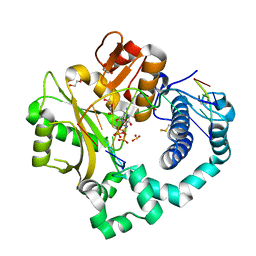 | | DNA Polymerase Mu, dGTP:Ct Reaction State Ternary Complex, 10 mM Mg2+ (10min) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KSY
 
 | | DNA Polymerase Mu, dGTP:Ct Product State Ternary Complex, 10 mM Mg2+ (960min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.577 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KSV
 
 | | DNA Polymerase Mu, dGTP:Ct Product State Ternary Complex, 10 mM Mn2+ (960min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KST
 
 | | DNA Polymerase Mu, dGTP:Ct Reaction State Ternary Complex, 10 mM Mn2+ (2min) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KT0
 
 | | DNA Polymerase Mu, dGTP:At Ground State Ternary Complex, 50 mM Mg2+ (60min) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
7KT1
 
 | | DNA Polymerase Mu, dGTP:At Reaction State Ternary Complex, 50 mM Mn2+ (180min) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-11-24 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.
Nat Commun, 12, 2021
|
|
