3WZK
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH N-cyclopropyl-4-(8-((thiophen-2-ylmethyl)amino)imidazo[1,2-a]pyrazin-3-yl)benzamide | | Descriptor: | CHLORIDE ION, Dual specificity protein kinase TTK, N-cyclopropyl-4-{8-[(thiophen-2-ylmethyl)amino]imidazo[1,2-a]pyrazin-3-yl}benzamide | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of imidazo[1,2-b]pyridazine derivatives: selective and orally available Mps1 (TTK) kinase inhibitors exhibiting remarkable antiproliferative activity.
J.Med.Chem., 58, 2015
|
|
2KUI
 
 | |
3ZRF
 
 | | pVHL54-213-EloB-EloC complex_apo | | Descriptor: | TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 2, VON HIPPEL-LINDAU DISEASE TUMOR SUPPRESSOR, | | Authors: | Van Molle, I, Buckley, D.L, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-06-16 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Von Hippel-Lindau E3 Ubiquitin Ligase Using Small Molecules to Disrupt the Vhl/Hif-1Alpha Interaction
J.Am.Chem.Soc., 134, 2012
|
|
6UJR
 
 | | P-glycoprotein mutant-F724A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
3ZV6
 
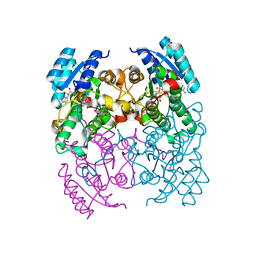 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB) FROM PANDORAEA PNOMENUSA STRAIN B-356 COMPLEX WITH CO-ENZYME NAD AND PRODUCT ANALOG 4,4'-DIHYDROXYBIPHENYL | | Descriptor: | 4,4'-DIHYDROXYBIPHENYL, CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
6UJT
 
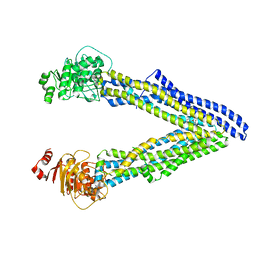 | | P-glycoprotein mutant-Y303A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
6FHQ
 
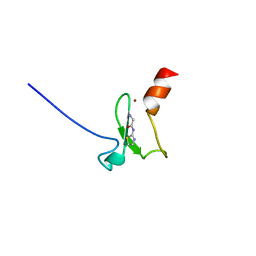 | | Crystal structure of human BAZ2B PHD zinc finger in complex with Fr 21 | | Descriptor: | 2-azanyl-~{N}-(1,3-thiazol-2-yl)ethanamide, Bromodomain adjacent to zinc finger domain protein 2B, ZINC ION | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6FHU
 
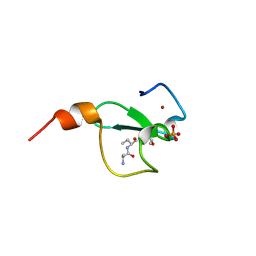 | | Crystal structure of BAZ2A PHD zinc finger in complex with H3 3-mer peptide | | Descriptor: | ALA-ARG-TAM, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-03-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6FC7
 
 | |
2KUD
 
 | |
2KUF
 
 | |
6UJW
 
 | | P-glycoprotein mutant-Y306A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
5OAX
 
 | | Galectin-3c in complex with thiogalactoside derivate | | Descriptor: | 5,6-bis(fluoranyl)-3-[[(2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-4-yl]oxymethyl]chromen-2-one, Galectin-3 | | Authors: | Nilsson, U.J, Peterson, K, Hakansson, M, Logan, D.T. | | Deposit date: | 2017-06-25 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Systematic Tuning of Fluoro-galectin-3 Interactions Provides Thiodigalactoside Derivatives with Single-Digit nM Affinity and High Selectivity.
J. Med. Chem., 61, 2018
|
|
6UIQ
 
 | |
6DHI
 
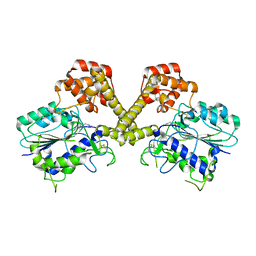 | |
8QD1
 
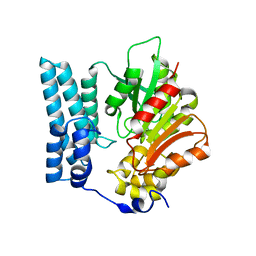 | | Ayg1p from A. fumigatus catalyzes polyketide shortening in the biosynthesis of DHN-melanin | | Descriptor: | Pigment biosynthesis protein yellowish-green 1 | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
1IXN
 
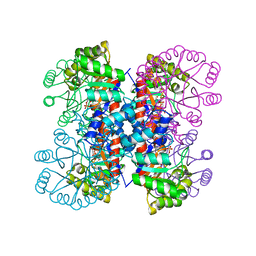 | | Enzyme-Substrate Complex of Pyridoxine 5'-Phosphate Synthase | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, Pyridoxine 5'-Phosphate Synthase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
4PFU
 
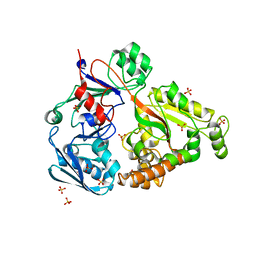 | | Crystal structure of mannobiose bound oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.05 A resolution | | Descriptor: | ABC transporter substrate-binding protein, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Lu, X, Ghimire-Rijal, S, Cuneo, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Duplication of Genes in an ATP-binding Cassette Transport System Increases Dynamic Range While Maintaining Ligand Specificity.
J.Biol.Chem., 289, 2014
|
|
5YVB
 
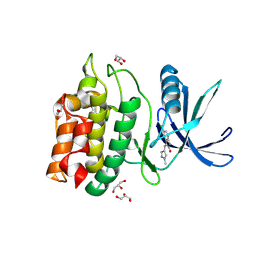 | | Structure of CaMKK2 in complex with CKI-011 | | Descriptor: | (3Z)-5-chloro-3-[(1-methyl-1H-pyrazol-4-yl)methylidene]-1,3-dihydro-2H-indol-2-one, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
5YV9
 
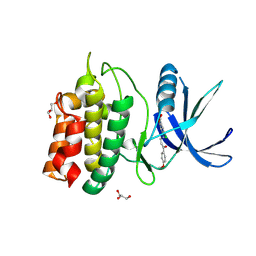 | | Structure of CaMKK2 in complex with CKI-009 | | Descriptor: | 5-chloro-2-methoxy-4[(1Z)-3-(4-methoxyphenyl)-3-oxoprop-1-en-1-yl]aminobenzoic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
7L4Z
 
 | | Structure of SARS-CoV-2 spike RBD in complex with cyclic peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-DTY-LYS-ALA-GLY-VAL-VAL-TYR-GLY-TYR-ASN-ALA-TRP-ILE-ARG-CYS-NH2, Spike protein S1 | | Authors: | Christie, M, Mackay, J.P, Passioura, T, Payne, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Discovery of Cyclic Peptide Ligands to the SARS-CoV-2 Spike Protein Using mRNA Display.
Acs Cent.Sci., 7, 2021
|
|
3ZV5
 
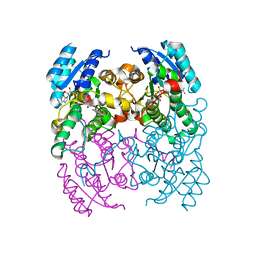 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB) FROM PANDORAEA PNOMENUSA STRAIN B-356 COMPLEX WITH CO-ENZYME NAD AND PRODUCT 2,3-DIHYDROXYBIPHENYL | | Descriptor: | BIPHENYL-2,3-DIOL, CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
6A1T
 
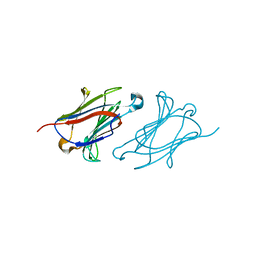 | | Charcot-Leyden crystal protein/Galectin-10 variant E33A with lactose | | Descriptor: | Galectin-10, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
3PGF
 
 | | Crystal structure of maltose bound MBP with a conformationally specific synthetic antigen binder (sAB) | | Descriptor: | GLYCEROL, IMIDAZOLE, Maltose-binding periplasmic protein, ... | | Authors: | Kossiakoff, A.A, Duguid, E.M, Sandstrom, A. | | Deposit date: | 2010-11-01 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
