5JO0
 
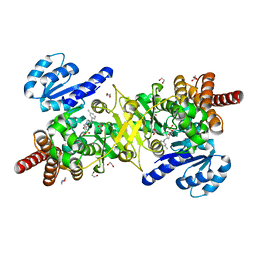 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC56 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
8WL6
 
 | | Structure of 1,3-beta-glucan synthase component FKS1 | | Descriptor: | 1,3-beta-glucan synthase component FKS1 | | Authors: | Li, J.L, Zhu, A.Q, Liu, J.X, Dai, X.L, Wang, X, Yan, C.Y, Deng, D. | | Deposit date: | 2023-09-29 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structure of the beta-1,3-glucan synthase FKS1-Rho1 complex.
Nat Commun, 16, 2025
|
|
6HAK
 
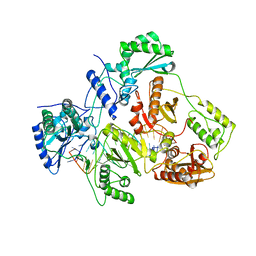 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with a double stranded RNA represents the RT transcription initiation complex prior to nucleotide incorporation | | Descriptor: | Gag-Pol polyprotein, MAGNESIUM ION, RNA (5'-R(P*AP*GP*UP*GP*GP*CP*GP*GP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2018-08-07 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of HIV-1 RT/dsRNA initiation complex prior to nucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6NKJ
 
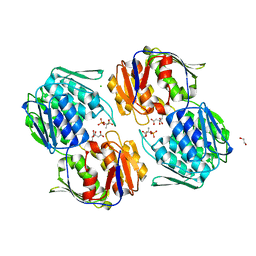 | | 1.3 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with (2R)-2-(phosphonooxy)propanoic acid. | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-07 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with (2R)-2-(phosphonooxy)propanoic acid.
To Be Published
|
|
6HMG
 
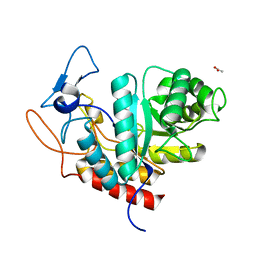 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-glucosamine) | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Lu, D, Zhu, S, Bernardo-Seisdedos, G, Millet, O, Zhang, Y, Sollogoub, M, Jimenez-Barbero, J, Davies, G.J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | From 1,4-Disaccharide to 1,3-Glycosyl Carbasugar: Synthesis of a Bespoke Inhibitor of Family GH99 Endo-alpha-mannosidase.
Org.Lett., 20, 2018
|
|
5XN2
 
 | | HIV-1 reverse transcriptase Q151M:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 38-MER DNA aptamer, GLYCEROL, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5JC1
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC55 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-14 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
5XN0
 
 | | HIV-1 reverse transcriptase Q151M:DNA binary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, Pol protein, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
6ONH
 
 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-IV-031-A05. | | Descriptor: | (3S)-N~3~-(4-chloro-3-fluorophenyl)-N~1~-propylpiperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
7APQ
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-(benzo[d]thiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-(1,3-benzothiazol-6-ylsulfonyl)-5-(methoxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Kolos, M.J, Pomplun, S, Riess, B, Purder, P, Voll, M.A, Merz, S, Bracher, A, Meyners, C, Krewald, V, Hausch, F. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Picomolar FKBP inhibitors enabled by a single water-displacing methyl group in bicyclic [4.3.1] aza-amides.
Chem Sci, 12, 2021
|
|
6OOO
 
 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-IV-226. | | Descriptor: | (3S,5R)-5-amino-N~3~-(4-chloro-3-fluorophenyl)-N~1~-propylpiperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-23 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of Small Molecules That Sensitize HIV-1 Infected Cells to Antibody-Dependent Cellular Cytotoxicity.
Acs Med.Chem.Lett., 11, 2020
|
|
5XN1
 
 | | HIV-1 reverse transcriptase Q151M:DNA:entecavir-triphosphate ternary complex | | Descriptor: | 38-MER DNA aptamer, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
8E8V
 
 | |
5JMP
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC57 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-29 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
6ZBM
 
 | | Structure of the D125N mutant of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6-alpha-manno-cyclophellitol carbasugar-stabilised trisaccharide inhibitor | | Descriptor: | (1~{R},2~{R},3~{R},4~{S},5~{R})-4-[[(1~{S},2~{S},3~{S},4~{R},5~{R})-5-(hydroxymethyl)-2,3,4-tris(oxidanyl)cyclohexyl]oxymethyl]cyclohexane-1,2,3,5-tetrol, 1,2-ETHANEDIOL, Alpha-1,6-mannanase, ... | | Authors: | Davies, G.J, Offen, W.A. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
6ZBX
 
 | | Structure of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6- alpha-manno-cyclophellitol carbasugar-stabilised trisaccharide inhibitor | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, (1~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, ... | | Authors: | Schroeder, S, Offen, W.A, Jin, Y, De Boer, C, Enoterpi, J, Marino, L, van der Marel, G.A, Codee, J.D.C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
6S34
 
 | |
7LOW
 
 | | S-adenosylmethionine synthetase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tan, L.L, Jackson, C.J. | | Deposit date: | 2021-02-10 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
7LOZ
 
 | | S-adenosylmethionine synthetase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tan, L.L, Jackson, C.J. | | Deposit date: | 2021-02-11 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
9EOV
 
 | | Crystal structure of domains I and II from the outer membrane cytochrome MtrC | | Descriptor: | 1,2-ETHANEDIOL, Extracellular iron oxide respiratory system surface decaheme cytochrome c component MtrC, HEME C, ... | | Authors: | Nash, B.W, Morales Florez, A, Lockwood, C.W.J, Edwards, M.J, Butt, J.N, Clarke, T.A. | | Deposit date: | 2024-03-15 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Extracellular catalysis of environmental substrates by Shewanella oneidensis MR-1 occurs via active sites on the C-terminal domains of MtrC.
Protein Sci., 34, 2025
|
|
6RXC
 
 | | Leishmania major pteridine reductase 1 (LmPTR1) in complex with inhibitor 4 (NMT-C0026) | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1, methyl 1-[4-[[2,4-bis(azanyl)pteridin-6-yl]methyl-(3-oxidanylpropyl)amino]phenyl]carbonylpiperidine-4-carboxylate | | Authors: | Di Pisa, F, Dello Iacono, L, Pozzi, C, Mangani, S. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multitarget, Selective Compound Design Yields Potent Inhibitors of a Kinetoplastid Pteridine Reductase 1.
J.Med.Chem., 65, 2022
|
|
7A9G
 
 | | Truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Mycobacterium tuberculosis with intermediate 2-acetyl-thiamine diphosphate | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, 2-ACETYL-THIAMINE DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-09-02 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First crystal structures of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Mycobacterium tuberculosis indicate a distinct mechanism of intermediate stabilization.
Sci Rep, 12, 2022
|
|
5JBI
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC52 and manganese | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-13 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
5J89
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-{2-[({2-methoxy-6-[(2-methyl[1,1'-biphenyl]-3-yl)methoxy]pyridin-3-yl}methyl)amino]ethyl}acetamide, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Guzik, K, Zieba, B.J, Musielak, B, Doemling, P, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1).
Oncotarget, 7, 2016
|
|
8WLA
 
 | | Cryo-EM structure of the beta-1,3-glucan synthase FKS1-Rho1 complex | | Descriptor: | 1,3-beta-glucan synthase component FKS1, GTP-binding protein RHO1 | | Authors: | Li, J.L, Zhu, A.Q, Liu, J.X, Dai, X.L, Yan, C.Y, Deng, D, Wang, X. | | Deposit date: | 2023-09-29 | | Release date: | 2025-02-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the beta-1,3-glucan synthase FKS1-Rho1 complex.
Nat Commun, 16, 2025
|
|
