6AAY
 
 | | the Cas13b binary complex | | Descriptor: | Bergeyella zoohelcum Cas13b (R1177A) mutant, RNA (52-MER) | | Authors: | Bo, Z, Weiwei, Y, Yangmiao, Y, Ouyang, S.Y. | | Deposit date: | 2018-07-19 | | Release date: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into Cas13b-guided CRISPR RNA maturation and recognition.
Cell Res., 28, 2018
|
|
8QOH
 
 | | Crystal structure of the kinetoplastid kinetochore protein KKT14 C-terminal domain from Apiculatamorpha spiralis | | Descriptor: | kinetochore protein KKT14 | | Authors: | Carter, W, Ballmer, D, Ishii, M, Ludzia, P, Akiyoshi, B. | | Deposit date: | 2023-09-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins.
Open Biology, 14, 2024
|
|
3VFP
 
 | | crystal structure of HLA B*3508 LPEP158G, HLA mutant Gly158 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, LPEP peptide from EBV, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
6S6Q
 
 | | Crystal structure of the LRR ectodomain of the plant membrane receptor kinase GASSHO1/SCHENGEN3 from Arabidopsis thaliana in complex with CASPARIAN STRIP INTEGRITY FACTOR 2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LRR receptor-like serine/threonine-protein kinase GSO1, ... | | Authors: | Okuda, S, Moretti, A, Hothorn, M. | | Deposit date: | 2019-07-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular mechanism for the recognition of sequence-divergent CIF peptides by the plant receptor kinases GSO1/SGN3 and GSO2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3O1G
 
 | |
6OO9
 
 | | Human CYP3A4 bound to a drug mibefradil | | Descriptor: | (1S,2S)-2-(2-{[3-(1H-benzimidazol-2-yl)propyl](methyl)amino}ethyl)-6-fluoro-1-(propan-2-yl)-1,2,3,4-tetrahydronaphthalen-2-yl methoxyacetate, 1,2-ETHANEDIOL, Cytochrome P450 3A4, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into the Interaction of Cytochrome P450 3A4 with Suicide Substrates: Mibefradil, Azamulin and 6',7'-Dihydroxybergamottin.
Int J Mol Sci, 20, 2019
|
|
7AXA
 
 | | Crystal structure of the hPXR-LBD in complex with clotrimazole | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Delfosse, V, Granell, M, Blanc, P, Bourguet, W. | | Deposit date: | 2020-11-09 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanistic insights into the synergistic activation of the RXR-PXR heterodimer by endocrine disruptor mixtures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7AXF
 
 | |
6OTS
 
 | | Rat ERK2 E320K | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Taylor, C.A, Cormier, K.W, Juang, Y.-C, Goldsmith, E.J, Cobb, M.H. | | Deposit date: | 2019-05-03 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence caused by mutations in an energetic hotspot in ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P0P
 
 | | Human beta-tryptase co-crystal structure with 5-{4-[3-(aminomethyl)phenyl]piperidine-1-carbonyl}-2-(3'-{4-[3-(aminomethyl)phenyl]piperidine-1-carbonyl}-[1,1'-biphenyl]-3-yl)-2-hydroxy-2H-1,3,2-benzodioxaborol-2-uide | | Descriptor: | (3'-{4-[3-(aminomethyl)phenyl]piperidine-1-carbonyl}[1,1'-biphenyl]-3-yl){4-[3-(aminomethyl)phenyl]piperidin-1-yl}[3,4-di(hydroxy-kappaO)phenyl]methanonato(2-)hydroxyborate(1-), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Giardina, S.F, Pingle, M.R, Werner, D.S, Feinberg, P.B, Foreman, K.W, Bergstrom, D.E, Arnold, L.D, Barany, F. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Novel, Self-Assembling Dimeric Inhibitors of Human beta Tryptase.
J.Med.Chem., 63, 2020
|
|
6AWT
 
 | | Structure of PR 10 Allergen in complex with epicatechin | | Descriptor: | (2R,3R)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol, BENZOIC ACID, PR 10 protein, ... | | Authors: | Offermann, L.R, Perdue, M, McBride, J, Hurlburt, B.K, Maleki, S.J, Pote, S.S, Chruszcz, M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of PR-10 Allergen Ara h 8.01.
To Be Published
|
|
6AVU
 
 | | Human alpha-V beta-3 Integrin (open conformation) in complex with the therapeutic antibody LM609 | | Descriptor: | Fab LM609 heavy chain, Fab LM609 light chain, Integrin alpha-V, ... | | Authors: | Borst, A.J, James, Z.N, Zagotta, W.N, Ginsberg, M, Rey, F.A, DiMaio, F, Backovic, M, Veesler, D. | | Deposit date: | 2017-09-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human alpha V beta 3 Integrin via Steric Hindrance.
Structure, 25, 2017
|
|
7BWI
 
 | | Solution structure of recombinant APETx1 | | Descriptor: | Kappa-actitoxin-Ael2a | | Authors: | Matsumura, K, Kobayashi, N, Kurita, J, Nishimura, Y, Yokogawa, M, Imai, S, Shimada, I, Osawa, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-23 | | Last modified: | 2021-07-14 | | Method: | SOLUTION NMR | | Cite: | Mechanism of hERG inhibition by gating-modifier toxin, APETx1, deduced by functional characterization.
Bmc Mol Cell Biol, 22, 2021
|
|
7BE6
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with inhibitor SR159 | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-N-(4-(((2S)-4-cyclohexyl-1-((1-(methylsulfonyl)piperidin-3-yl)amino)-1-oxobutan-2-yl)carbamoyl)benzyl)-1-phenyl-1H-pyrazole-4-carboxamide, Epithelial discoidin domain-containing receptor 1, ... | | Authors: | Pinkas, D.M, Bufton, J.C, Roehm, S, Joerger, A.C, Knapp, S, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87081933 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
3OM3
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1SM1
 
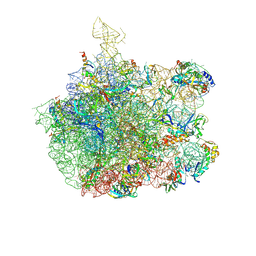 | | COMPLEX OF THE LARGE RIBOSOMAL SUBUNIT FROM DEINOCOCCUS RADIODURANS WITH QUINUPRISTIN AND DALFOPRISTIN | | Descriptor: | 23S RIBOSOMAL RNA, 5-(2-DIETHYLAMINO-ETHANESULFONYL)-21-HYDROXY-10-ISOPROPYL-11,19-DIMETHYL-9,26-DIOXA-3,15,28-TRIAZA-TRICYCLO[23.2.1.00,255]OCTACOSA-1(27),12,17,19,25(28)-PENTAENE-2,8,14,23-TETRAONE, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Harms, J.M, Schluenzen, F, Fucini, P, Bartels, H, Yonath, A. | | Deposit date: | 2004-03-08 | | Release date: | 2004-08-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Alterations at the Peptidyl Transferase Centre of the Ribosome Induced by the Synergistic Action of the Streptogramins Dalfopristin and Quinupristin.
Bmc Biol., 2, 2004
|
|
1A4K
 
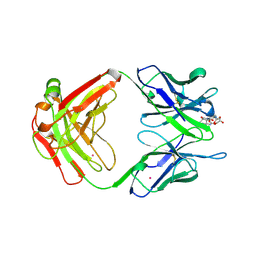 | | DIELS ALDER CATALYTIC ANTIBODY WITH TRANSITION STATE ANALOGUE | | Descriptor: | ANTIBODY FAB, CADMIUM ION, [4-(4-ACETYLAMINO-PHENYL)-3,5-DIOXO-4-AZA-TRICYCLO[5.2.2.0 2,6]UNDEC-1-YLCARBAMOYLOXY]-ACETIC ACID | | Authors: | Spiller, B.W, Romesburg, F.E, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Immunological origins of binding and catalysis in a Diels-Alderase antibody.
Science, 279, 1998
|
|
6BZP
 
 | | STGGYG from low-complexity domain of FUS, residues 77-82 | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, RNA-binding protein FUS | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-25 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
3OPY
 
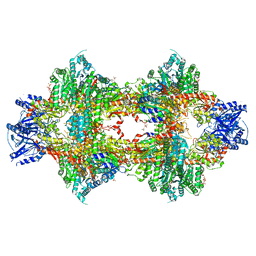 | | Crystal structure of Pichia pastoris phosphofructokinase in the T-state | | Descriptor: | 6-phosphofructo-1-kinase alpha-subunit, 6-phosphofructo-1-kinase beta-subunit, 6-phosphofructo-1-kinase gamma-subunit, ... | | Authors: | Strater, N, Marek, S, Kuettner, E.B, Kloos, M, Keim, A, Bruser, A, Kirchberger, J, Schoneberg, T. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular architecture and structural basis of allosteric regulation of eukaryotic phosphofructokinases.
Faseb J., 25, 2011
|
|
1RLD
 
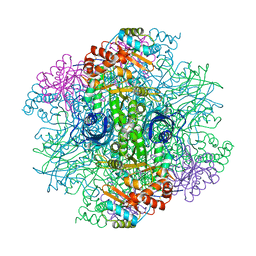 | | SOLID-STATE PHASE TRANSITION IN THE CRYSTAL STRUCTURE OF RIBULOSE 1,5-BIPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE | | Descriptor: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Eisenberg, D. | | Deposit date: | 1993-12-10 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solid-state phase transition in the crystal structure of ribulose 1,5-bisphosphate carboxylase/oxygenase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
6P7E
 
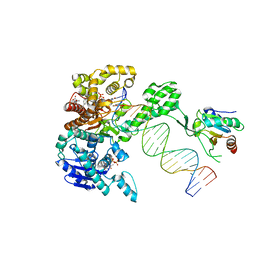 | | Structure of T7 DNA Polymerase Bound to a Primer/Template DNA and a Peptide that Mimics the C-terminal Tail of the Primase-Helicase | | Descriptor: | ASP-THR-ASP-PHE peptide, DNA (25-MER), DNA (5'-D(P*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*CP*TP*TP*GP*CP*CP*GP*GP*TP*GP*A)-3'), ... | | Authors: | Foster, B.M, Rosenberg, D, Salvo, H, Stephens, K.L, Bintz, B.J, Hammel, M, Ellenberger, T, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2019-06-05 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Combined Solution and Crystal Methods Reveal the Electrostatic Tethers That Provide a Flexible Platform for Replication Activities in the Bacteriophage T7 Replisome.
Biochemistry, 58, 2019
|
|
8T3W
 
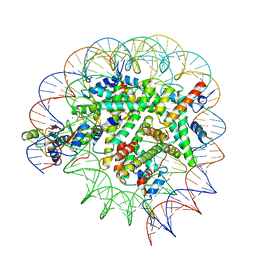 | | Structure of Bre1-nucleosome complex - state2 | | Descriptor: | 601 DNA strand 1, 601 DNA strand 2, E3 ubiquitin-protein ligase BRE1, ... | | Authors: | Zhao, F, Hicks, C.W, Wolberger, C. | | Deposit date: | 2023-06-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T3T
 
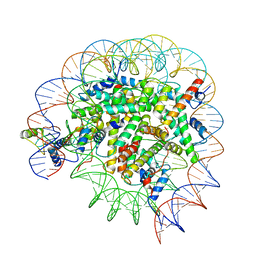 | | Structure of Bre1-nucleosome complex - state3 | | Descriptor: | 601 DNA strand 1, 601 DNA strand 2, E3 ubiquitin-protein ligase BRE1, ... | | Authors: | Zhao, F, Hicks, C.W, Wolberger, C. | | Deposit date: | 2023-06-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T3Y
 
 | | Structure of Bre1-nucleosome complex - state1 | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, E3 ubiquitin-protein ligase BRE1, ... | | Authors: | Zhao, F, Hicks, C.W, Wolberger, C. | | Deposit date: | 2023-06-08 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
