2WIG
 
 | | NONAGED FORM OF HUMAN BUTYRYLCHOLINESTERASE INHIBITED BY TABUN ANALOGUE TA4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Carletti, E, Aurbek, N, Gillon, E, Loiodice, M, Nicolet, Y, Fontecilla, J, Masson, P, Thiermann, H, Nachon, F, Worek, F. | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-activity analysis of aging and reactivation of human butyrylcholinesterase inhibited by analogues of tabun.
Biochem. J., 421, 2009
|
|
6E3Y
 
 | | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | | Descriptor: | Calcitonin gene-related peptide 1, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Deganutti, G, Glukhova, A, Koole, C, Peat, T.S, Radjainia, M, Plitzko, J.M, Baumeister, W, Miller, L.J, Hay, D.L, Christopoulos, A, Reynolds, C.A, Wootten, D, Sexton, P.M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor.
Nature, 561, 2018
|
|
2VYU
 
 | | CRYSTAL STRUCTURE OF CHOLINE BINDING PROTEIN F FROM STREPTOCOCCUS PNEUMONIAE IN THE PRESENCE OF A PEPTIDOGLYCAN ANALOGUE (TETRASACCHARIDE-PENTAPEPTIDE) | | Descriptor: | CHOLINE BINDING PROTEIN F, CHOLINE ION | | Authors: | Perez-Dorado, I, Molina, R, Hermoso, J.A, Mobashery, S. | | Deposit date: | 2008-07-28 | | Release date: | 2009-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Cbpf, a Bifunctional Choline-Binding Protein and Autolysis Regulator from Streptococcus Pneumoniae.
Embo Rep., 10, 2009
|
|
7CBQ
 
 | | Crystal structure of PDE4D catalytic domain in complex with Apremilast | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-{2-[(1S)-1-(3-ethoxy-4-methoxyphenyl)-2-(methylsulfonyl)ethyl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}acetamide, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
2WKH
 
 | | Crystal structure of the acyl-enzyme OXA-10 K70C-Ampicillin at pH 7 | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, BETA-LACTAMASE OXA-10, SULFATE ION | | Authors: | Vercheval, L, Bauvois, C, Kerff, F, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
7MBO
 
 | | FACTOR XIA (PICHIA PASTORIS; C500S [C122S]) IN COMPLEX WITH THE INHIBITOR Milvexian (BMS-986177), IUPAC NAME:(6R,10S)-10-{4-[5-chloro-2-(4-chloro-1H-1,2,3-triazol-1-yl)phenyl]-6- oxopyrimidin-1(6H)-yl}-1-(difluoromethyl)-6-methyl-1,4,7,8,9,10-hexahydro-15,11- (metheno)pyrazolo[4,3-b][1,7]diazacyclotetradecin-5(6H)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, Milvexian | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.924 Å) | | Cite: | Discovery of Milvexian, a High-Affinity, Orally Bioavailable Inhibitor of Factor XIa in Clinical Studies for Antithrombotic Therapy.
J.Med.Chem., 65, 2022
|
|
5OY4
 
 | | GSK3beta complex with N-(6-(3,4-dihydroxyphenyl)-1H-pyrazolo[3,4-b]pyridin-3-yl)acetamide | | Descriptor: | Glycogen synthase kinase-3 beta, Proto-oncogene FRAT1, SULFATE ION, ... | | Authors: | Bax, B.D, Convery, M.A. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | From PIM1 to PI3K delta via GSK3 beta : Target Hopping through the Kinome.
ACS Med Chem Lett, 8, 2017
|
|
6U2M
 
 | | Crystal structure of a HaloTag-based calcium indicator, HaloCaMP V2, bound to JF635 | | Descriptor: | (1E,3S)-1-{10-[2-carboxy-5-({2-[2-(hexyloxy)ethoxy]ethyl}carbamoyl)phenyl]-7-(3-fluoroazetidin-1-yl)-5,5-dimethyldibenz o[b,e]silin-3(5H)-ylidene}-3-fluoroazetidin-1-ium, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Deo, C, Schreiter, E.R. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HaloTag as a general scaffold for far-red tunable chemigenetic indicators.
Nat.Chem.Biol., 17, 2021
|
|
3GON
 
 | | Streptococcus pneumoniae Phosphomevalonate Kinase in Complex with Phosphomevalonate and AMPPNP | | Descriptor: | (3R)-3-HYDROXY-3-METHYL-5-(PHOSPHONOOXY)PENTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Andreassi, J.L, Bilder, P.W, Vetting, M.W, Roderick, S.L, Leyh, T.S. | | Deposit date: | 2009-03-19 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the ternary complex of phosphomevalonate kinase: the enzyme and its family
Biochemistry, 48, 2009
|
|
3H9J
 
 | | Crystal structure of E. coli MccB + AMPCPP + SeMeT MccA | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MccB protein, Microcin C7 ANALOG, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
6O74
 
 | |
8CPB
 
 | | 1,6-anhydro-n-actetylmuramic acid kinase (AnmK) in complex with AMPPNP, and AnhMurNAc at 1.7 Angstroms resolution. | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, GLYCEROL, ... | | Authors: | Jimenez-Faraco, E, Hermoso, J.A. | | Deposit date: | 2023-03-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic process of anhydro-N-acetylmuramic acid kinase from Pseudomonas aeruginosa.
J.Biol.Chem., 299, 2023
|
|
1TX7
 
 | | Bovine Trypsin complexed with p-amidinophenylmethylphosphinic acid (AMPA) | | Descriptor: | (4-CARBAMIMIDOYLPHENYL)-METHYL-PHOSPHINIC ACID, CALCIUM ION, Trypsinogen | | Authors: | Cui, J, Marankan, F, Fu, W, Crich, D, Mesecar, A, Johnson, M.E. | | Deposit date: | 2004-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An oxyanion-hole selective serine protease inhibitor in complex with trypsin.
Bioorg.Med.Chem., 10, 2002
|
|
6BXE
 
 | |
6TCH
 
 | |
6CI7
 
 | |
6ZSN
 
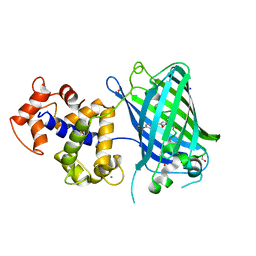 | | Crystal structure of rsGCaMP double mutant Ile80His/Val116Ile in the OFF state (illuminated) | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Green fluorescent protein,Calmodulin, ... | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-07-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 40, 2022
|
|
6ZSM
 
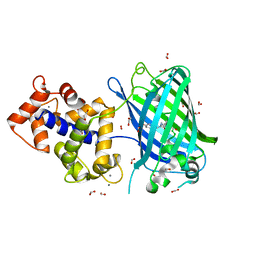 | | Crystal structure of rsGCaMP double mutant Ile80His/Val116Ile in the ON state (non-illuminated) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FORMIC ACID, ... | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-07-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 40, 2022
|
|
6I0I
 
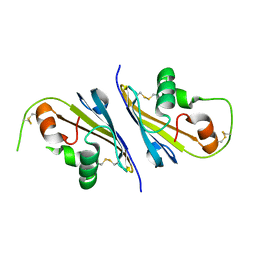 | |
3HUN
 
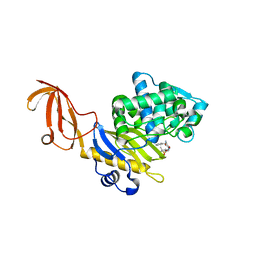 | |
8DVR
 
 | | Cryo-EM structure of RIG-I bound to the end of p3SLR30 (+AMPPNP) | | Descriptor: | Antiviral innate immune response receptor RIG-I, GUANOSINE-5'-TRIPHOSPHATE, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
6WQV
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose | | Descriptor: | 1,2-ETHANEDIOL, Endoglucanase, NITRATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
3TB9
 
 | | Structure of Yeast Ribonucleotide Reductase 1 Q288A with AMPPNP and CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-08-05 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Role of Arginine 293 and Glutamine 288 in Communication between Catalytic and Allosteric Sites in Yeast Ribonucleotide Reductase.
J.Mol.Biol., 419, 2012
|
|
6C2R
 
 | | Aurora A ligand complex | | Descriptor: | (2R,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-4-({3-fluoro-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyridin-2-yl}methyl)-2-methylpiperidine-4-carboxylic acid, Aurora kinase A, SULFATE ION | | Authors: | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
6C2T
 
 | | Aurora A ligand complex | | Descriptor: | (2S,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-2-methyl-4-({3-[(1,3-thiazol-2-yl)amino]isoquinolin-1-yl}methyl)piperidine-4-carboxylic acid, Aurora kinase A, DIMETHYL SULFOXIDE, ... | | Authors: | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
