4R1R
 
 | |
7DW9
 
 | |
4REL
 
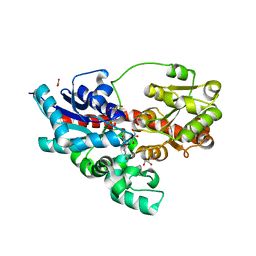 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with kaempferol | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ACETATE ION, GLYCEROL, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
7DTY
 
 | | Structural basis of ligand selectivity conferred by the human glucose-dependent insulinotropic polypeptide receptor | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhang, C, Zhou, Q.T, Hang, K.N, Zou, X.Y, Chen, Y, Wu, F, Rao, Q.D, Dai, A.T, Yin, W.C, Shen, D.D, Zhang, Y, Xia, T, Stevens, R.C, Xu, H.E, Yang, D.H, Zhao, L.H, Wang, M.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into hormone recognition by the human glucose-dependent insulinotropic polypeptide receptor.
Elife, 10, 2021
|
|
4RNQ
 
 | | Crystal structure of tobacco 5-epi-aristolochene synthase (TEAS) with anilinogeranyl diphosphate (AGPP) and geraniline | | Descriptor: | (2E,6E)-3,7-dimethyl-8-(phenylamino)octa-2,6-dien-1-yl trihydrogen diphosphate, 5-epi-aristolochene synthase, ACETATE ION, ... | | Authors: | Koo, H.J, Crenshaw, C.M, Starks, C, Spielmann, H.P, Chappell, J, Noel, J.P. | | Deposit date: | 2014-10-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Formation of a Novel Macrocyclic Alkaloid from the Unnatural Farnesyl Diphosphate Analogue Anilinogeranyl Diphosphate by 5-Epi-Aristolochene Synthase.
Acs Chem.Biol., 10, 2015
|
|
7DUR
 
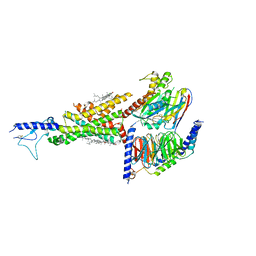 | | Cryo-EM structure of the compound 2-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-08-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
4TQB
 
 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2E)-2-{2-[4-(4-bromophenyl)-1,3-thiazol-2-yl]hydrazinylidene}-3-(2-nitrophenyl)propanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5TJ8
 
 | |
4TUH
 
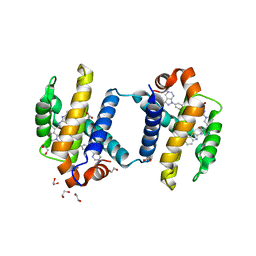 | | Bcl-xL in complex with inhibitor (Compound 10) | | Descriptor: | 1,2-ETHANEDIOL, 2-[8-(1,3-benzothiazol-2-ylcarbamoyl)-3,4-dihydroisoquinolin-2(1H)-yl]-5-{3-[4-(1H-pyrazolo[3,4-d]pyrimidin-1-yl)phenoxy]propyl}-1,3-thiazole-4-carboxylic acid, ACETATE ION, ... | | Authors: | Czabotar, P.E, Lessense, G, Smith, B.J, Colman, P.M. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Rescaffolding of Selective Antagonists of BCL-XL.
Acs Med.Chem.Lett., 5, 2014
|
|
7XJI
 
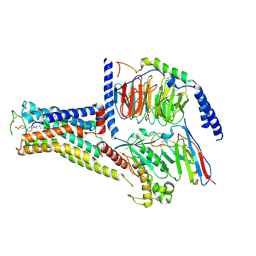 | | Solabegron-activated dog beta3 adrenergic receptor | | Descriptor: | 3-[3-[2-[[(2~{S})-2-(3-chlorophenyl)-2-oxidanyl-ethyl]amino]ethylamino]phenyl]benzoic acid, Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the beta 3 adrenergic receptor bound to solabegron and isoproterenol.
Biochem.Biophys.Res.Commun., 611, 2022
|
|
7DB6
 
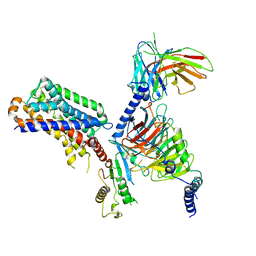 | | human melatonin receptor MT1 - Gi1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Okamoto, H.H, Kusakizako, T, Shihioya, W, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-10-19 | | Release date: | 2021-08-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human MT 1 -G i signaling complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2H13
 
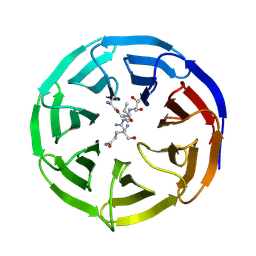 | |
7XJH
 
 | | Isoproterenol-activated dog beta3 adrenergic receptor | | Descriptor: | Beta-3 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shihoya, W, Nureki, O. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the beta 3-adrenergic receptor reveals the molecular basis of subtype selectivity.
Mol.Cell, 81, 2021
|
|
7Y35
 
 | | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex | | Descriptor: | Abaloparatide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex
To Be Published
|
|
2GV2
 
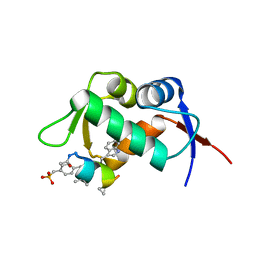 | | MDM2 in complex with an 8-mer p53 peptide analogue | | Descriptor: | 8-MER P53 PEPTIDE ANALOGUE, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Schubert, C, Sakurai, K. | | Deposit date: | 2006-05-02 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Analysis of an 8-mer p53 Peptide Analogue Complexed with MDM2.
J.Am.Chem.Soc., 128, 2006
|
|
7XO4
 
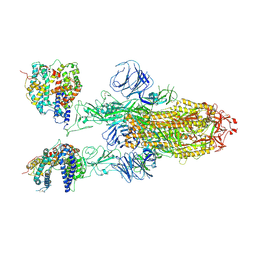 | | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-04-30 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO6
 
 | | SARS-CoV-2 Omicron BA.1 Variant RBD with mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOA
 
 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with one mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
4RVB
 
 | | Crystal Structure Analysis of the Human Leukotriene A4 Hydrolase | | Descriptor: | ACETATE ION, ACETIC ACID, GLYCEROL, ... | | Authors: | Ouyang, P, Lu, W, Cui, K, Huang, J. | | Deposit date: | 2014-11-25 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure Analysis of the Human Leukotriene A4 Hydrolase
To be Published
|
|
7XOC
 
 | | SARS-CoV-2 Omicron BA.2 Variant RBD complexed with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOB
 
 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with two mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO5
 
 | | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with one mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XZ5
 
 | | GPR119-Gs-LPC complex | | Descriptor: | (4R,7R,18E)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XJL
 
 | | Cryo-EM structure of the spexin-bound GALR2-miniGq complex | | Descriptor: | Galanin receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, W, Zheng, S. | | Deposit date: | 2022-04-18 | | Release date: | 2022-06-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into galanin receptor signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XZ6
 
 | | GPR119-Gs-APD668 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
