7XZQ
 
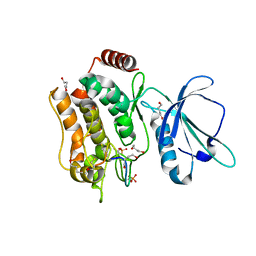 | | Crystal structure of TNIK-thiopeptide TP1 complex | | Descriptor: | 1,4-BUTANEDIOL, TRAF2 and NCK-interacting protein kinase, thiopeptide TP1 | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
5IJJ
 
 | |
9LJ3
 
 | | Crystal structure of P25 | | Descriptor: | 3-[[4-chloranyl-2-[(2-hydroxyethylamino)methyl]-5-[[2-methyl-3-[3-[2-[2-[(3~{R})-3-oxidanylpyrrolidin-1-yl]ethoxy]ethoxy]phenyl]phenyl]methoxy]phenoxy]methyl]benzenecarbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Yang, P, Chen, M.R, Xiao, Y.B. | | Deposit date: | 2025-01-14 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery and Crystallography Study of Novel Resorcinol Dibenzyl Ether-Based PD-1/PD-L1 Inhibitors with Improved Drug-like and Pharmacokinetic Properties for Cancer Treatment.
J.Med.Chem., 68, 2025
|
|
6DUB
 
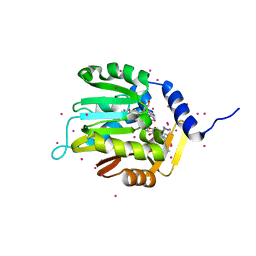 | | Crystal structure of a methyltransferase | | Descriptor: | Alpha N-terminal protein methyltransferase 1B, GLYCEROL, RCC1, ... | | Authors: | Dong, C, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An asparagine/glycine switch governs product specificity of human N-terminal methyltransferase NTMT2.
Commun Biol, 1, 2018
|
|
6T8Y
 
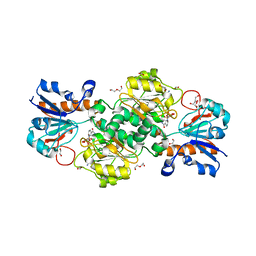 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
9KXM
 
 | | Crystal structure of the PIN1 and fragment 57 complex. | | Descriptor: | (1~{S})-1-(1~{H}-benzimidazol-2-yl)ethanol, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Xiao, Q.J, Wu, T.T, Shu, H.L, Qin, W.M. | | Deposit date: | 2024-12-06 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Uncovering druggable hotspots on Pin1 via X-ray crystallographic fragment screening.
Eur.J.Med.Chem., 299, 2025
|
|
9HQ6
 
 | | Structural insights in the HuHf@gold-monocarbene adduct: aurophilicity revealed in a biological context | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, Ferritin heavy chain, GOLD ION | | Authors: | Cosottini, L, Giachetti, A, Guerri, A, Martinez-Castillo, A, Geri, A, Zineddu, S, Abrescia, N.G.A, Messori, L, Turano, P, Rosato, A. | | Deposit date: | 2024-12-16 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (1.51 Å) | | Cite: | Structural Insight Into a Human H Ferritin@Gold-Monocarbene Adduct: Aurophilicity Revealed in a Biological Context.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6PXP
 
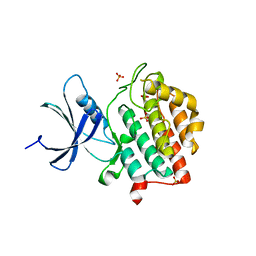 | | Human Casein Kinase 1 delta Site 2 mutant (K171E) | | Descriptor: | Casein kinase I isoform delta, SULFATE ION | | Authors: | Yee, L, Philpott, J.M, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2019-07-26 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Casein kinase 1 dynamics underlie substrate selectivity and the PER2 circadian phosphoswitch.
Elife, 9, 2020
|
|
5K0R
 
 | | Crystal structure of reduced Shewanella Yellow Enzyme 4 (SYE4) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H:flavin oxidoreductase Sye4, Octadecane | | Authors: | Elegheert, J, Brige, A, Savvides, S.N. | | Deposit date: | 2016-05-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural dissection of Shewanella oneidensis old yellow enzyme 4 bound to a Meisenheimer complex and (nitro)phenolic ligands.
FEBS Lett., 591, 2017
|
|
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Chen, S.J, Ye, F. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
5GZ3
 
 | | Structure of D-amino acid dehydrogenase in complex with NADP | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | Deposit date: | 2016-09-26 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
6QAN
 
 | |
8XGL
 
 | | Structure of the Magnaporthe oryzae effector NIS1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Necrosis-inducing secreted protein 1, ... | | Authors: | Han, R, Wang, D, Zhang, X, Liu, J. | | Deposit date: | 2023-12-15 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Understanding and manipulating the recognition of necrosis-inducing secreted protein 1 (NIS1) by BRI1-associated receptor kinase 1 (BAK1).
Int.J.Biol.Macromol., 278, 2024
|
|
8DV3
 
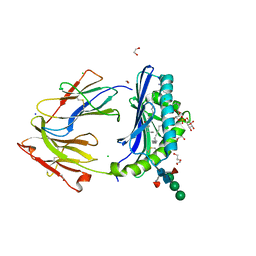 | | Crystal structure of human CD1b presenting Phosphatidylinositol C34:1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Farquhar, R, Rossjohn, J, Shahine, A. | | Deposit date: | 2022-07-28 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | alpha beta T-cell receptor recognition of self-phosphatidylinositol presented by CD1b.
J.Biol.Chem., 299, 2023
|
|
9EY4
 
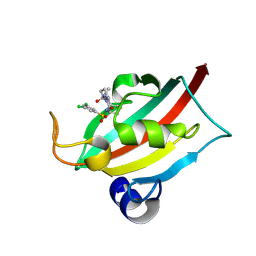 | | The FK1 domain of FKBP51 in complex with (3S,11S)-12-((3,5-dichlorophenyl)sulfonyl)-5-oxo-11-vinyldecahydro-1H-6,10-epiminopyrrolo[1,2-a]azonine-3-carboxamide | | Descriptor: | (1~{S},4~{S},7~{S},8~{S},9~{R})-13-[3,5-bis(chloranyl)phenyl]sulfonyl-8-ethenyl-2-oxidanylidene-3,13-diazatricyclo[7.3.1.0^{3,7}]tridecane-4-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Krajczy, P, Hausch, F. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure-Based Design of Ultrapotent Tricyclic Ligands for FK506-Binding Proteins.
Chemistry, 30, 2024
|
|
9EY3
 
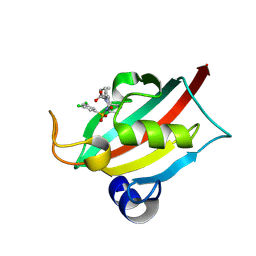 | | The FK1 domain of FKBP51 in complex with (3S,11S,11aS)-12-((3,5-dichlorophenyl)sulfonyl)-5-oxo-11-vinyldecahydro-1H-6,10-epiminopyrrolo[1,2-a]azonine-3-carboxylic acid | | Descriptor: | (1~{S},4~{S},7~{S},8~{S},9~{R})-13-[3,5-bis(chloranyl)phenyl]sulfonyl-8-ethenyl-2-oxidanylidene-3,13-diazatricyclo[7.3.1.0^{3,7}]tridecane-4-carboxylic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Krajczy, P, Hausch, F. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure-Based Design of Ultrapotent Tricyclic Ligands for FK506-Binding Proteins.
Chemistry, 30, 2024
|
|
6I4Q
 
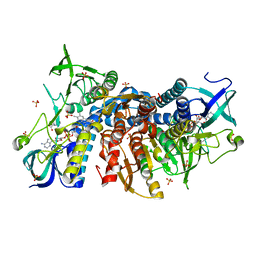 | | Crystal structure of the human dihydrolipoamide dehydrogenase at 1.75 Angstrom resolution | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Nagy, B, Szabo, E, Wilk, P, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
5K8K
 
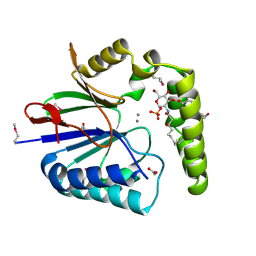 | | Structure of the Haemophilus influenzae LpxH-lipid X complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, ACETATE ION, GLYCEROL, ... | | Authors: | Cho, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the essential Haemophilus influenzae UDP-diacylglucosamine pyrophosphohydrolase LpxH in lipid A biosynthesis.
Nat Microbiol, 1, 2016
|
|
5KA6
 
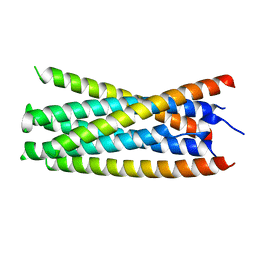 | |
5Y52
 
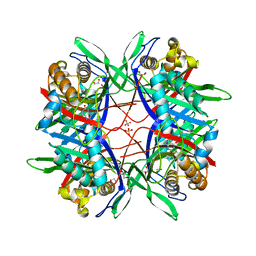 | |
7ZZF
 
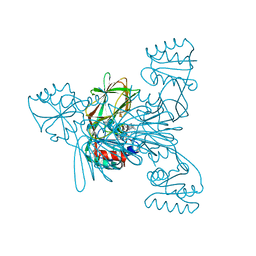 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},23~{R},24~{S},25~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-24,25-bis(oxidanyl)-26-oxa-2,4,6,9,14,17,21-heptazatetracyclo[21.2.1.0^{2,10}.0^{3,8}]hexacosa-3(8),4,6,9-tetraen-11-yne-18,20-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
7ZZE
 
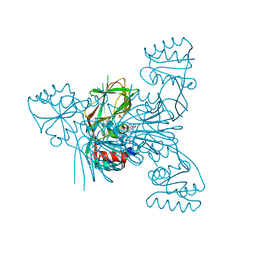 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},25~{R},26~{S},27~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-26,27-bis(oxidanyl)-28-oxa-2,4,6,9,14,17,21,23-octazatetracyclo[23.2.1.0^{2,10}.0^{3,8}]octacosa-3(8),4,6,9-tetraen-11-yne-16,22-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
5GUV
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - R1279D | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Ye, F, Chen, S.J. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.078 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
4ZHE
 
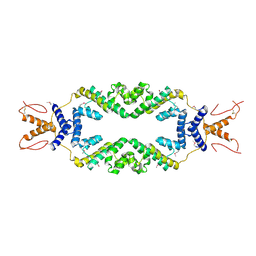 | | Crystal structure of the SeMet substituted Topless related protein 2 (TPR2) N-terminal domain (1-209) from rice | | Descriptor: | ASPR2 protein | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors.
Sci Adv, 1, 2015
|
|
6UAQ
 
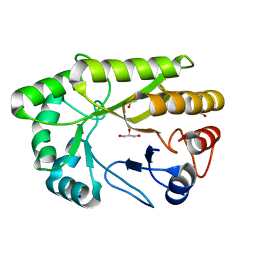 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) | | Descriptor: | GLYCEROL, Glyco_hydro_cc domain-containing protein, SODIUM ION | | Authors: | Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
