3H8X
 
 | | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked host-guest system | | Descriptor: | 5'-D(*CP*TP*GP*TP*(ME6)P*TP*(2YR)P*AP*TP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*AP*TP*AP*AP*GP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2 | | Authors: | Lu, L, Yi, C, Jian, X, Zheng, G, He, C. | | Deposit date: | 2009-04-29 | | Release date: | 2010-03-31 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system.
Nucleic Acids Res., 38, 2010
|
|
4AAE
 
 | | Crystal structure of the mutant D75N I-CreI in complex with an altered target (The four central bases, 2NN region, are composed by AGCG from 5' to 3') | | Descriptor: | 24MER DNA, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AAB
 
 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 10MER DNA 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP*GP*AP)-3', 14MER DNA 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*CP)-3', DNA ENDONUCLEASE I-CREI, ... | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AAD
 
 | | Crystal structure of the mutant D75N I-CreI in complex with its wild- type target in absence of metal ions at the active site (The four central bases, 2NN region, are composed by GTAC from 5' to 3') | | Descriptor: | 24MER DNA, DNA ENDONUCLEASE I-CREI, GLYCEROL | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
4AAF
 
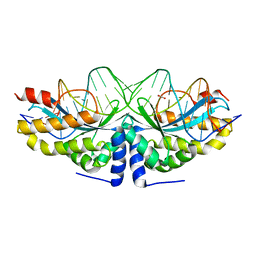 | | Crystal structure of the mutant D75N I-CreI in complex with an altered target (The four central bases, 2NN region, are composed by TGCA from 5' to 3') | | Descriptor: | 1,2-ETHANEDIOL, 24MER DNA, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
2GUI
 
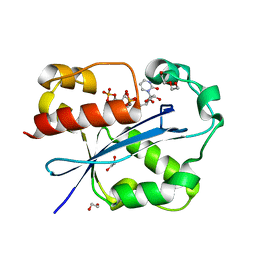 | | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit of E. coli DNA Polymerase III | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA polymerase III epsilon subunit, ... | | Authors: | Park, A.Y, Carr, P.D, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2006-04-30 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of Cyclized Versions of the Proofreading Exonuclease Subunit E. coli DNA Polymerase III
To be Published
|
|
2FDJ
 
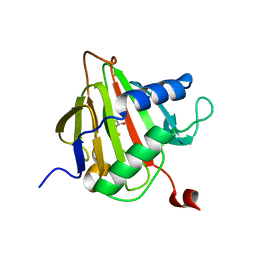 | | Crystal Structure of AlkB in complex with Fe(II) and succinate | | Descriptor: | Alkylated DNA repair protein alkB, FE (II) ION, SUCCINIC ACID | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2AY0
 
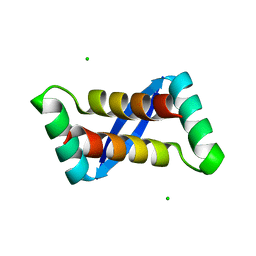 | | Structure of the Lys9Met mutant of the E. coli Proline Utilization A (PutA) DNA-binding domain. | | Descriptor: | Bifunctional putA protein, CHLORIDE ION | | Authors: | Larson, J.D, Schuermann, J.P, Zhou, Y, Jenkins, J.L, Becker, D.F, Tanner, J.J. | | Deposit date: | 2005-09-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition.
Protein Sci., 15, 2006
|
|
2L4L
 
 | | Structural insights into the cTAR DNA recognition by the HIV-1 Nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC | | Descriptor: | 5'-D(*CP*TP*GP*G)-3', HIV-1 nucleocapsid protein NCp7, ZINC ION | | Authors: | Bazzi, A, Zargarian, L, Chaminade, F, Boudier, C, De Rocquigny, H, Rene, B, Mely, Y, Fosse, P, Mauffret, O. | | Deposit date: | 2010-10-08 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC.
Nucleic Acids Res., 39, 2011
|
|
1IDX
 
 | |
2IDO
 
 | | Structure of the E. coli Pol III epsilon-Hot proofreading complex | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III epsilon subunit, Hot protein, ... | | Authors: | Kirby, T.W, Harvey, S, DeRose, E.F, Chalov, S, Chikova, A.K, Perrino, F.W, Schaaper, R.M, London, R.E, Pedersen, L.C. | | Deposit date: | 2006-09-15 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Escherichia coli DNA polymerase III epsilon-HOT proofreading complex.
J.Biol.Chem., 281, 2006
|
|
5BQT
 
 | | Structure of TrmBL2, an archaeal chromatin protein, shows a novel mode of DNA binding. | | Descriptor: | CALCIUM ION, Putative HTH-type transcriptional regulator TrmBL2 | | Authors: | Ahmad, M.U, Diederichs, K, Welte, W. | | Deposit date: | 2015-05-29 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into Nonspecific Binding of DNA by TrmBL2, an Archaeal Chromatin Protein.
J.Mol.Biol., 427, 2015
|
|
3NX5
 
 | | The crystal structure of Sanguinarine bound to DNA d(CGTACG) | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, 5'-D(*CP*GP*TP*AP*CP*G)-3', CALCIUM ION | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | Deposit date: | 2010-07-13 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | X-Ray diffraction analyses of the natural isoquinoline alkaloids Berberine and Sanguinarine complexed with double helix DNA d(CGTACG)
Chem.Commun.(Camb.), 47, 2011
|
|
1XC9
 
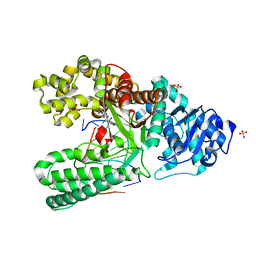 | | Structure of a high-fidelity polymerase bound to a benzo[a]pyrene adduct that blocks replication | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Huang, X, Luneva, N.P, Geacintov, N.E, Beese, L.S. | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a High Fidelity DNA Polymerase Bound to a Benzo[a]pyrene Adduct That Blocks Replication
J.Biol.Chem., 280, 2005
|
|
3A1B
 
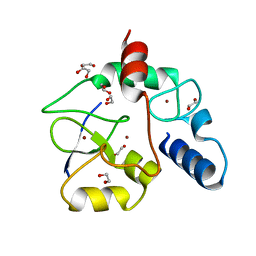 | | Crystal structure of the DNMT3A ADD domain in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 3A, Histone H3.1, ... | | Authors: | Otani, J, Arita, K, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2009-03-28 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain
Embo Rep., 10, 2009
|
|
2VBJ
 
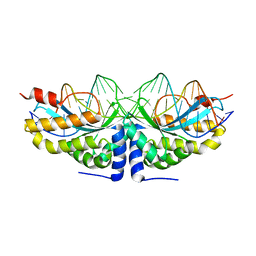 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*CP*TP*TP*TP*TP *TP*TP*GP*AP*AP*GP*GP*AP*TP*CP*CP*TP*AP*A)-3', 5'-D(*TP*TP*AP*GP*GP*AP*TP*CP*CP*TP *TP*CP*AP*AP*AP*AP*AP*AP*GP*GP*CP*AP*GP*A)-3', CALCIUM ION, ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
7JTQ
 
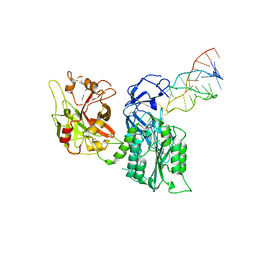 | |
7JTN
 
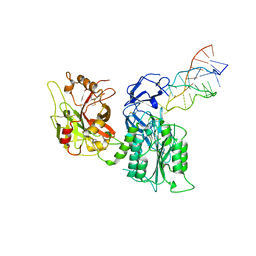 | |
6HPH
 
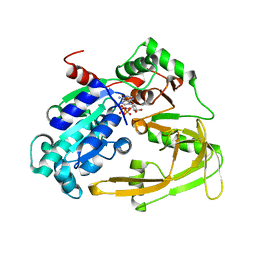 | | Crystal structure of human Pif1 helicase in complex with AMP-PNP | | Descriptor: | ATP-dependent DNA helicase PIF1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Levdikov, V.M, Dehghani-Tafti, S, Bax, B.D, Sanders, C.M, Antson, A.A. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural and functional analysis of the nucleotide and DNA binding activities of the human PIF1 helicase.
Nucleic Acids Res., 47, 2019
|
|
6HPU
 
 | | Crystal structure of human Pif1 helicase in complex with ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, MAGNESIUM ION, ... | | Authors: | Levdikov, V.M, Dehghani-Tafti, S, Bax, B.D, Sanders, C.M, Antson, A.A. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Structural and functional analysis of the nucleotide and DNA binding activities of the human PIF1 helicase.
Nucleic Acids Res., 47, 2019
|
|
7K72
 
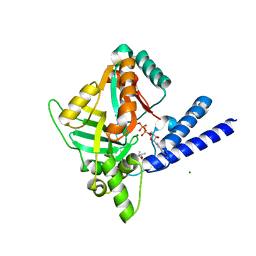 | |
8CZQ
 
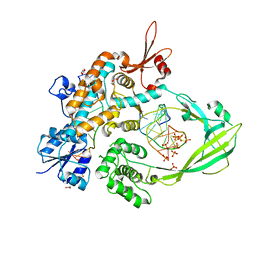 | | The crystal structure of MtbTOP1 in complex with both G- and T-segments | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*TP*TP*CP*CP*GP*CP*TP*TP*GP*AP*C)-3'), DNA topoisomerase 1, ... | | Authors: | Tan, K, Tse-Dinh, Y.-C. | | Deposit date: | 2022-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The interaction between transport-segment DNA and topoisomerase IA-crystal structure of MtbTOP1 in complex with both G- and T-segments.
Nucleic Acids Res., 51, 2023
|
|
6HPT
 
 | | Crystal structure of human Pif1 helicase, apoform. | | Descriptor: | ATP-dependent DNA helicase PIF1, SULFATE ION | | Authors: | Levdikov, V.M, Dehghani-Tafti, S, Bax, B.D, Sanders, C.M, Antson, A.A. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural and functional analysis of the nucleotide and DNA binding activities of the human PIF1 helicase.
Nucleic Acids Res., 47, 2019
|
|
2VE9
 
 | | Xray structure of KOPS bound gamma domain of FtsK (P. aeruginosa) | | Descriptor: | 5'-D(*AP*CP*CP*AP*GP*GP*GP*CP*AP*GP *GP*GP*CP*GP*AP*C)-3', 5'-D(*GP*TP*CP*GP*CP*CP*CP*TP*GP*CP *CP*CP*TP*GP*GP*T)-3', DNA TRANSLOCASE FTSK, ... | | Authors: | Lowe, J, Allen, M.D, Sherratt, D.J. | | Deposit date: | 2007-10-17 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Mechanism of Sequence-Directed DNA Loading and Translocation by Ftsk.
Mol.Cell, 31, 2008
|
|
2VAH
 
 | | Solution structure of a B-DNA hairpin at low pressure. | | Descriptor: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | Authors: | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Change in a B-DNA Helix with Hydrostatic Pressure
Nucleic Acids Res., 36, 2008
|
|
