6IZ3
 
 | |
5Z9C
 
 | |
2IX9
 
 | |
2IZV
 
 | | CRYSTAL STRUCTURE OF SOCS-4 IN COMPLEX WITH ELONGIN-B AND ELONGIN-C AT 2.55A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Debreczeni, J.E, Bullock, A, Papagrigoriou, E, Turnbull, A, Pike, A.C.W, Gorrec, F, von Delft, F, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Knapp, S. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the SOCS4-ElonginB/C complex reveals a distinct SOCS box interface and the molecular basis for SOCS-dependent EGFR degradation.
Structure, 15, 2007
|
|
6KD0
 
 | | Crystal Structure of Vibralactone Cyclase | | Descriptor: | Vibralactone Cyclase | | Authors: | Zeng, Y, Feng, K.N. | | Deposit date: | 2019-06-30 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Hydrolase-Catalyzed Cyclization Forms the Fused Bicyclic beta-Lactone in Vibralactone.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2IVX
 
 | | Crystal structure of human cyclin T2 at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CYCLIN-T2 | | Authors: | Debreczeni, J.E, Bullock, A.N, Fedorov, O, Savitsky, P, Berridge, G, Das, S, Pike, A.C.W, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Gorrec, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation.
EMBO J., 27, 2008
|
|
2IXA
 
 | | A-zyme, N-acetylgalactosaminidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-N-ACETYLGALACTOSAMINIDASE, ... | | Authors: | Sulzenbacher, G, Liu, Q.P, Bourne, Y, Henrissat, B, Clausen, H. | | Deposit date: | 2006-07-07 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial Glycosidases for the Production of Universal Red Blood Cells.
Nat.Biotechnol., 25, 2007
|
|
2IXB
 
 | | Crystal structure of N-ACETYLGALACTOSAMINIDASE in complex with GalNAC | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Sulzenbacher, G, Liu, Q.P, Bourne, Y, Henrissat, B, Clausen, H. | | Deposit date: | 2006-07-07 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacterial Glycosidases for the Production of Universal Red Blood Cells.
Nat.Biotechnol., 25, 2007
|
|
4P3C
 
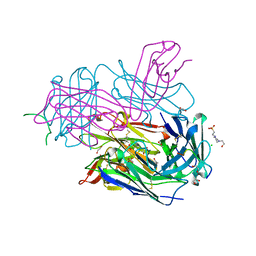 | | MT1-MMP:Fab complex (Form I) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Rozenberg, H, Udi, Y, Sagi, I. | | Deposit date: | 2014-03-06 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
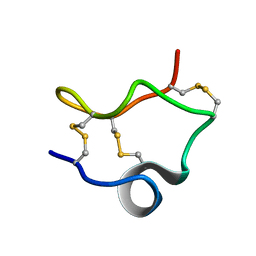
2LIX
 
 | |
2NDF
 
 | |
4P3D
 
 | | MT1-MMP:Fab complex (Form II) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Rozenberg, H, Udi, Y, Sagi, I. | | Deposit date: | 2014-03-07 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
2NDG
 
 | |
3HU9
 
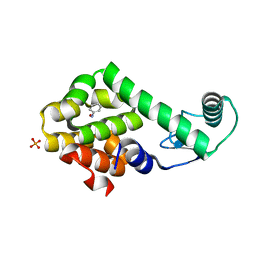 | | Nitrosobenzene in complex with T4 lysozyme L99A/M102Q | | Descriptor: | Lysozyme, NITROSOBENZENE, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
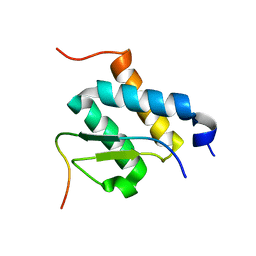
2ND0
 
 | |
2NCZ
 
 | |
2ND1
 
 | |
5MLA
 
 | | Crystal structure of human RAS in complex with darpin K55 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Overman, R, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Inhibition of RAS nucleotide exchange by a DARPin: structural characterisation and effects on downstream signalling by active RAS
To Be Published
|
|
6CBY
 
 | | Crystal structure of human SET and MYND Domain Containing protein 2 with MTF9975 | | Descriptor: | N-lysine methyltransferase SMYD2, ZINC ION, [3-(4-amino-6-methyl-1H-imidazo[4,5-c]pyridin-1-yl)-3-methylazetidin-1-yl][1-({1-[(1R)-cyclohept-2-en-1-yl]piperidin-4-yl}methyl)-1H-pyrrol-3-yl]methanone | | Authors: | ZENG, H, DONG, A, Hutchinson, A, Seitova, A, TATLOCK, J, KUMPF, R, OWEN, A, TAYLOR, A, Casimiro-Garcia, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-05 | | Release date: | 2018-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Selective, Small-Molecule Co-Factor Binding Site Inhibition of a Su(var)3-9, Enhancer of Zeste, Trithorax Domain Containing Lysine Methyltransferase.
J.Med.Chem., 62, 2019
|
|
6ZNC
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human wild-type p53DBD bound to DNA and MQ: wt-DNA-MQ (I) | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Degtjarik, O, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2020-07-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
7B4F
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R282W mutant bound to DNA: R282W-MQ (I) | | Descriptor: | Cellular tumor antigen p53, DNA target, FORMIC ACID, ... | | Authors: | Rozenberg, H, Degtjarik, O, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
7B4D
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R273C/S240R double mutant bound to DNA and MQ: R273C/S240R-DNA-MQ | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Diskin-Posner, Y, Degtjarik, O, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
7B4N
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human wild-type p53DBD bound to DNA and MQ: wt-DNA-MQ (II) | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Diskin-Posner, Y, Degtjarik, O, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
7B49
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R273H mutant bound to DNA and MQ: R273H-DNA-MQ | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, 1,2-ETHANEDIOL, ... | | Authors: | Rozenberg, H, Degtjarik, O, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
5O2S
 
 | | Human KRAS in complex with darpin K27 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange.
Nat Commun, 8, 2017
|
|
