4R80
 
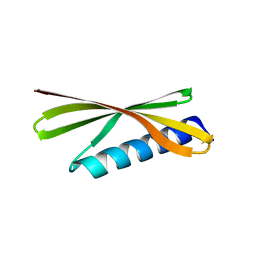 | | Crystal Structure of a De Novo Designed Beta Sheet Protein, Cystatin Fold, Northeast Structural Genomics Consortium (NESG) Target OR486 | | Descriptor: | OR486 | | Authors: | Guan, R, Marcos, E, O'Connell, P, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Crystal Structure of an engineered protein with denovo beta sheet design, Northeast Structural Genomics Consortium (NESG) Target OR486
To be Published
|
|
6GFW
 
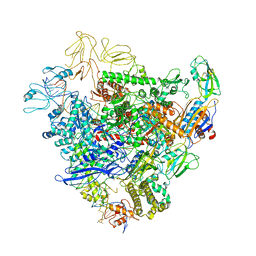 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme initial transcribing complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
8P4Y
 
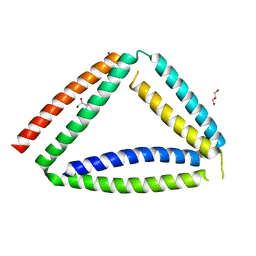 | | Coiled-coil protein origami triangle | | Descriptor: | GLYCEROL, Protein origami triangle | | Authors: | Satler, T, Hadzi, S, Jerala, R. | | Deposit date: | 2023-05-23 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal Structure of de Novo Designed Coiled-Coil Protein Origami Triangle.
J.Am.Chem.Soc., 145, 2023
|
|
1D3G
 
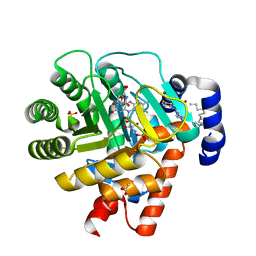 | | HUMAN DIHYDROOROTATE DEHYDROGENASE COMPLEXED WITH BREQUINAR ANALOG | | Descriptor: | 2-BIPHENYL-4-YL-6-FLUORO-3-METHYL-QUINOLINE-4-CARBOXYLIC ACID, ACETATE ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Liu, S, Neidhardt, E.A, Grossman, T.H, Ocain, T, Clardy, J. | | Deposit date: | 1999-09-29 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents.
Structure Fold.Des., 8, 2000
|
|
1D3H
 
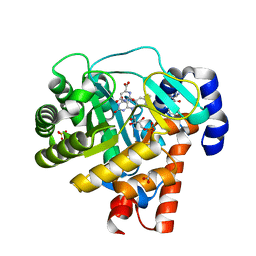 | | HUMAN DIHYDROOROTATE DEHYDROGENASE COMPLEXED WITH ANTIPROLIFERATIVE AGENT A771726 | | Descriptor: | (2Z)-2-cyano-3-hydroxy-N-[4-(trifluoromethyl)phenyl]but-2-enamide, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Liu, S, Neidhardt, E.A, Grossman, T.H, Ocain, T, Clardy, J. | | Deposit date: | 1999-09-29 | | Release date: | 2000-08-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents.
Structure Fold.Des., 8, 2000
|
|
7N1J
 
 | |
6M6Z
 
 | | A de novo designed transmembrane nanopore, TMH4C4 | | Descriptor: | TMH4C4 | | Authors: | Lu, P, Xu, C, Reggiano, G, Xu, Q, DiMaio, F, Baker, D. | | Deposit date: | 2020-03-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
1QP6
 
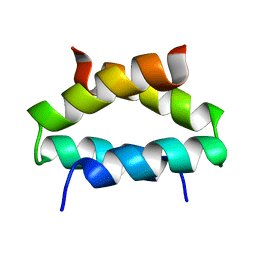 | | SOLUTION STRUCTURE OF ALPHA2D | | Descriptor: | PROTEIN (ALPHA2D) | | Authors: | Hill, R.B, DeGrado, W.F. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Alpha2D, A Nativelike De Novo Designed Protein
J.Am.Chem.Soc., 120, 1998
|
|
1I4U
 
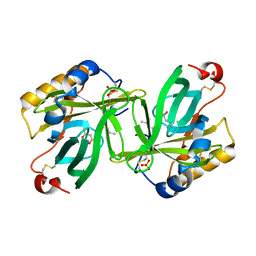 | | THE C1 SUBUNIT OF ALPHA-CRUSTACYANIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CRUSTACYANIN, SULFATE ION | | Authors: | Gordon, E.J, Leonard, G.A, McSweeney, S, Zagalsky, P.F. | | Deposit date: | 2001-02-23 | | Release date: | 2001-09-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The C1 subunit of alpha-crustacyanin: the de novo phasing of the crystal structure of a 40 kDa homodimeric protein using the anomalous scattering from S atoms combined with direct methods.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7DNS
 
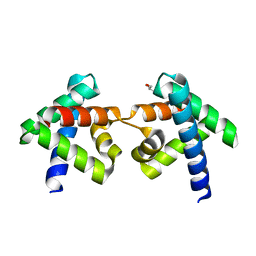 | | Crystal structure of domain-swapped dimer of H5_Fold-0 Elsa; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | GLYCEROL, de novo designed protein | | Authors: | Suzuki, K, Kobayashi, N, Murata, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-12-10 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7DMF
 
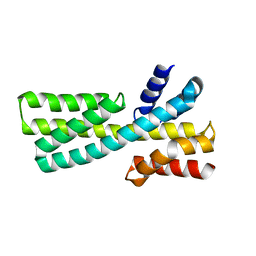 | |
7WCQ
 
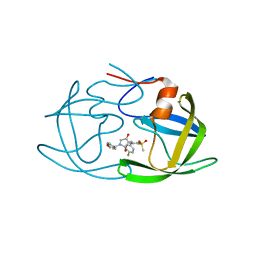 | | Crystal structure of HIV-1 protease in complex with lactam derivative 1 | | Descriptor: | (3R,4R)-3-[(4-fluorophenyl)methyl]-1-[(4-methoxyphenyl)methyl]-3-(4-methylsulfonylphenyl)-4-oxidanyl-pyrrolidin-2-one, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakasa-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
7WBS
 
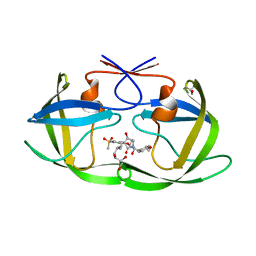 | | Crystal structure of HIV-1 protease in complex with lactam derivative 2 | | Descriptor: | (3~{R},4~{R})-1-[(4-methoxyphenyl)methyl]-3-(3-methylbutyl)-3-[4-methylsulfonyl-2-[(2~{S})-1-oxidanylpropan-2-yl]oxy-phenyl]-4-oxidanyl-pyrrolidin-2-one, GLYCEROL, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakabayashi-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | Authors: | Rao, X, Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5LKA
 
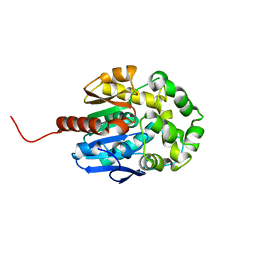 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 at 1.3 A resolution | | Descriptor: | Haloalkane dehalogenase, THIOCYANATE ION | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2016-07-21 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Engineering a de novo transport tunnel.
Acs Catalysis, 2016
|
|
7LDF
 
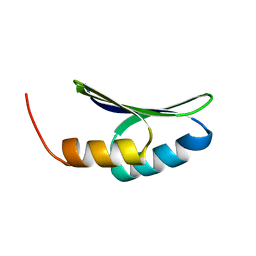 | |
5LY1
 
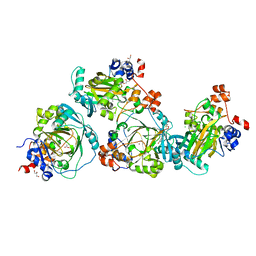 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II) AND Macrocyclic PEPTIDE Inhibitor CP2 (13-mer) | | Descriptor: | CHLORIDE ION, CP2, GLYCEROL, ... | | Authors: | King, O.N.F, Chowdhury, R, Kawamura, A, Schofield, C.J. | | Deposit date: | 2016-09-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Nat Commun, 8, 2017
|
|
5LY2
 
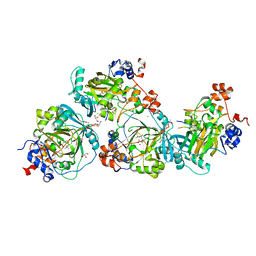 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II), NOG AND Macrocyclic PEPTIDE Inhibitor CP2_R6Kme3 (13-mer) | | Descriptor: | CHLORIDE ION, CP2_R6Kme3, GLYCEROL, ... | | Authors: | Chowdhury, R, Madden, S.K, Hopkinson, R, Schofield, C.J. | | Deposit date: | 2016-09-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Nat Commun, 8, 2017
|
|
3A7E
 
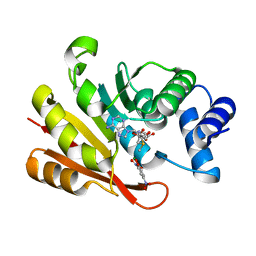 | | Crystal structure of human COMT complexed with SAM and 3,5-dinitrocatechol | | Descriptor: | 3,5-DINITROCATECHOL, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Tsuji, E. | | Deposit date: | 2009-09-26 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hit to Lead: Comprehensive Strategy of de novo Scaffold Generation by FBDD. Part 1: In silico Fragments Linking and Verification of Spatial Proximity using Inter Ligand NOE Approachs
To be Published
|
|
3A7D
 
 | | Crystal Structures of rat Catechol-O-Methyltransferase complexed with new bi-substrate type inhibitor | | Descriptor: | 5'-deoxy-5'-[4-({[(2,3-dihydroxy-5-nitrophenyl)carbonyl]amino}methyl)-1H-1,2,3-triazol-1-yl]adenosine, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Tsuji, E. | | Deposit date: | 2009-09-26 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hit to Lead: Comprehensive Strategy of de novo Scaffold Generation by FBDD. Part 2: Ligand Fishing using Mass Spectrometry by Detection of Ligand-Protein Non-Covalent Complex after Matrix Click Chemistry
To be Published
|
|
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
7UZL
 
 | |
