8BH1
 
 | | Core divisome complex FtsWIQBL from Pseudomonas aeruginosa | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Kaeshammer, L, van den Ent, F, Jeffery, M, Lowe, J. | | Deposit date: | 2022-10-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the bacterial divisome core complex and antibiotic target FtsWIQBL.
Nat Microbiol, 8, 2023
|
|
5U47
 
 | | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus | | Descriptor: | ACETATE ION, CHLORIDE ION, Penicillin binding protein 2X | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-03 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus.
To Be Published
|
|
6Q9N
 
 | | Crystal structure of PBP2a from MRSA in complex with piperacillin and quinazolinone | | Descriptor: | 3-[2-[(~{E})-2-(4-ethynylphenyl)ethenyl]-4-oxidanylidene-quinazolin-3-yl]benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Martinez-Caballero, S, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Quinazolinone Allosteric Inhibitor of PBP 2a Synergizes with Piperacillin and Tazobactam against Methicillin-Resistant Staphylococcus aureus.
Antimicrob.Agents Chemother., 63, 2019
|
|
4KQQ
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
8C5W
 
 | | Crystal Structure of Penicillin-binding Protein 3 (PBP3) from Staphylococcus Epidermidis in complex with Cefotaxime | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Schwinzer, M, Brognaro, H, Rohde, H, Betzel, C. | | Deposit date: | 2023-01-10 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure and Dynamics of the Penicillin-Binding Protein 3 from Staphylococcus Epidermidis Native and in Complex with Cefotaxime and Vaborbactam
Int J Appl Biol Pharm, 2023
|
|
8C5B
 
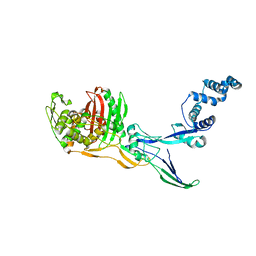 | |
8C5O
 
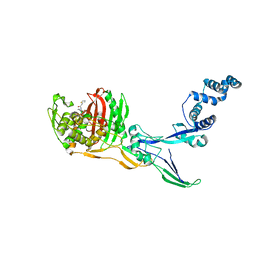 | |
4BL3
 
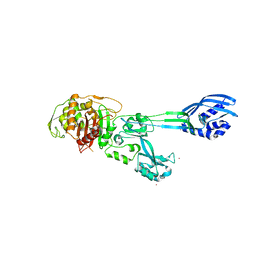 | | Crystal structure of PBP2a clinical mutant N146K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, PENICILLIN BINDING PROTEIN 2 PRIME, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Carrasco-Lopez, C, Hermoso, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
4KQR
 
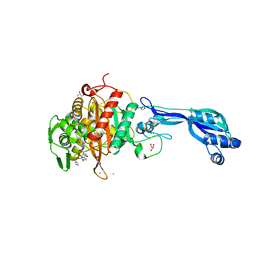 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4CPK
 
 | | Crystal structure of PBP2a double clinical mutant N146K-E150K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, Penicillin binding protein 2 prime, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
2ZC3
 
 | | Penicillin-binding protein 2X (PBP 2X) acyl-enzyme complex (biapenem) from Streptococcus pneumoniae | | Descriptor: | (4R,5S)-3-(6,7-dihydro-5H-pyrazolo[1,2-a][1,2,4]triazol-4-ium-6-ylsulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-meth yl-4,5-dihydro-1H-pyrrole-2-carboxylate, Penicillin-binding protein 2X, SULFATE ION | | Authors: | Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Biapenem and Tebipenem Complexed with Penicillin-Binding Proteins 2X and 1A from Streptococcus pneumoniae
Antimicrob.Agents Chemother., 52, 2008
|
|
6R40
 
 | |
2Z2L
 
 | |
4L0L
 
 | | Crystal structure of P.aeruginosa PBP3 in complex with compound 4 | | Descriptor: | (6R,7S,10Z)-10-(2-amino-1,3-thiazol-4-yl)-1-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-7-formyl-13,13-dimethyl-3,9-dioxo-6-(sulfoamino)-12-oxa-2,4,8,11-tetraazatetradec-10-en-14-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S, Marr, E.S. | | Deposit date: | 2013-05-31 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyridone-conjugated monobactam antibiotics with gram-negative activity.
J.Med.Chem., 56, 2013
|
|
5UY7
 
 | |
4KQO
 
 | | Crystal structure of penicillin-binding protein 3 from pseudomonas aeruginosa in complex with piperacillin | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4CJN
 
 | | Crystal structure of PBP2a from MRSA in complex with quinazolinone ligand | | Descriptor: | (E)-3-(2-(4-cyanostyryl)-4-oxoquinazolin-3(4H)-yl)benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Bouley, R, Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2013-12-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Discovery of Antibiotic (E)-3-(3-Carboxyphenyl)-2-(4-Cyanostyryl)Quinazolin-4(3H)-One.
J.Am.Chem.Soc., 137, 2015
|
|
2Z2M
 
 | | Cefditoren-Acylated Penicillin-Binding Protein 2X (PBP2X) from Streptococcus pneumoniae | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(2-AMINO-1,3-THIAZOL-4-YL)-2-(METHOXYIMINO)ACETYL]AMINO}-2-OXOETHYL]-5-[(Z)-2-(4-METHYL-1,3-THIAZOL-5-YL)VINYL]-3,6-DIHYDRO-2H-1,3-THIAZINE-4-CARBOXYLIC ACID, Penicillin-binding protein 2X, SULFATE ION | | Authors: | Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-05-23 | | Release date: | 2007-09-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Cefditoren Complexed with Streptococcus pneumoniae Penicillin-Binding Protein 2X: Structural Basis for its High Antimicrobial Activity
Antimicrob.Agents Chemother., 51, 2007
|
|
2ZC4
 
 | | Penicillin-binding protein 2X (PBP 2X) acyl-enzyme complex (tebipenem) from Streptococcus pneumoniae | | Descriptor: | (4R,5S)-3-(1-(4,5-dihydrothiazol-2-yl)azetidin-3-ylthio)-5-((2S,3R)-3-hydroxy-1-oxobutan-2-yl)-4-methyl-4,5- dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 2X, SULFATE ION | | Authors: | Yamada, M, Watanabe, T, Takeuchi, Y. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Biapenem and Tebipenem Complexed with Penicillin-Binding Proteins 2X and 1A from Streptococcus pneumoniae
Antimicrob.Agents Chemother., 52, 2008
|
|
8VBW
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Ertapenem) inhibited form | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBV
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Cephalexin) inhibited form | | Descriptor: | (2S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBT
 
 | |
8VBU
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Oxacillin) inhibited form | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
5M19
 
 | | Crystal structure of PBP2a from MRSA in the presence of Oxacillin ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
5LP5
 
 | | Complex between Penicillin-Binding Protein (PBP2) and MreC from Helicobacter pylori | | Descriptor: | Penicillin-binding protein 2 (Pbp2), Rod shape-determining protein (MreC) | | Authors: | Contreras-Martel, C, Martins, A, Ecobichon, C, Maragno, D.M, Mattei, P.J, El Ghachi, M, Hicham, S, Hardouin, P, Boneca, I.G, Dessen, A. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.
Nat Commun, 8, 2017
|
|
