6AW4
 
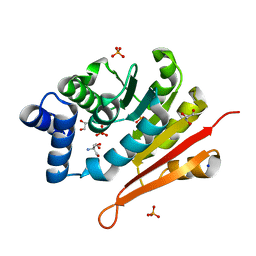 | | 1.50A resolution structure of catechol O-methyltransferase (COMT) from Nannospalax galili | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Catechol O-methyltransferase, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6YK4
 
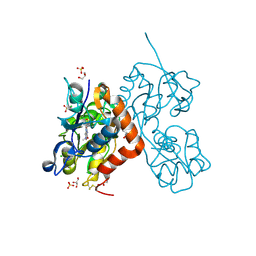 | | Structure of the AMPA receptor GluA2o ligand-binding domain (S1S2J) in complex with the compound ( S) - 1- [2'-Amino-2'-carboxyethyl]-6-methyl-5 ,7- dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 1.00A | | Descriptor: | (2~{S})-2-azanyl-3-[6-methyl-2,4-bis(oxidanylidene)-5,7-dihydropyrrolo[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2020-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | Ionotropic Glutamate Receptor GluA2 in Complex with Bicyclic Pyrimidinedione-Based Compounds: When Small Compound Modifications Have Distinct Effects on Binding Interactions.
Acs Chem Neurosci, 11, 2020
|
|
8A39
 
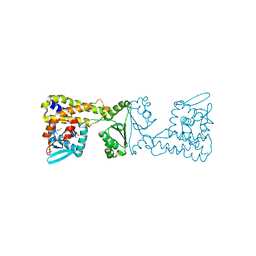 | | Crystal Structure of PaaX from Escherichia coli W | | Descriptor: | DNA-binding transcriptional repressor of phenylacetic acid degradation, aryl-CoA responsive, GLYCEROL, ... | | Authors: | Molina, R, Alba-Perez, A, Hermoso, J.A. | | Deposit date: | 2022-06-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of PaaX, the main repressor of the phenylacetate degradation pathway in Escherichia coli W: A novel fold of transcription regulator proteins.
Int.J.Biol.Macromol., 254, 2024
|
|
6YK2
 
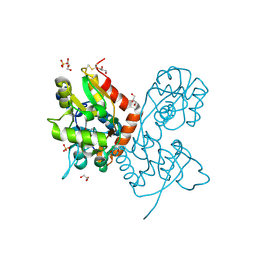 | | Structure of the AMPA receptor GluA2o ligand-binding domain (S1S2J) in complex with the compound (S)-1-[2'-Amino-2'-carboxyethyl]-5,7-dihydrothieno[3,4-d]pyrimidin- 2,4(1H,3H)-dione at resolution 1.60A | | Descriptor: | (2~{S})-2-azanyl-3-[2,4-bis(oxidanylidene)-5,7-dihydrothieno[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2020-04-05 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Ionotropic Glutamate Receptor GluA2 in Complex with Bicyclic Pyrimidinedione-Based Compounds: When Small Compound Modifications Have Distinct Effects on Binding Interactions.
Acs Chem Neurosci, 11, 2020
|
|
6BSW
 
 | | Crystal structure of Xyloglucan Xylosyltransferase 1 ternary form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Culbertson, A.T, Ehrlich, J.J, Choe, J, Honzatko, R.B, Zabotina, O.A. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-23 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7OPR
 
 | | Rab27a fusion with Slp2a-RBDa1 effector covalent adduct with CB1 in C123 | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
6D33
 
 | | Crystal structure of BH1352 2-deoxyribose-5-phosphate from Bacillus halodurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Kim, T, Yim, V, Yakunin, A, Savchenko, A. | | Deposit date: | 2018-04-14 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Rational engineering of 2-deoxyribose-5-phosphate aldolases for the biosynthesis of (R)-1,3-butanediol.
J.Biol.Chem., 295, 2020
|
|
4Z2E
 
 | | Quinolone(Trovafloxacin)-DNA cleavage complex of gyrase from S. pneumoniae | | Descriptor: | DNA gyrase subunit A, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Selvarajah, J, Crevel, I.M.-T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2015-03-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structural studies of the drug-stabilized cleavage complexes of topoisomerase IV and gyrase from Streptococcus pneumoniae
To Be Published
|
|
6GXJ
 
 | | X-ray structure of DiRu-1-encapsulated Apoferritin | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Pica, A, Ferraro, G, Merlino, A. | | Deposit date: | 2018-06-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Encapsulation of the Dinuclear Trithiolato-Bridged Arene Ruthenium Complex Diruthenium-1 in an Apoferritin Nanocage: Structure and Cytotoxicity.
ChemMedChem, 14, 2019
|
|
5I8L
 
 | | Crystal structure of the full-length cell wall-binding module of Cpl7 mutant R223A | | Descriptor: | GLYCEROL, Lysozyme | | Authors: | Bernardo-Garcia, N, Silva-Martin, N, Uson, I, Hermoso, J.A. | | Deposit date: | 2016-02-19 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Deciphering how Cpl-7 cell wall-binding repeats recognize the bacterial peptidoglycan.
Sci Rep, 7, 2017
|
|
6RHJ
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-(4-benzyl-1,4-diazepane-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-[[4-(phenylmethyl)-1,4-diazepan-1-yl]carbonyl]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
4Z0W
 
 | | Peptaibol gichigamin isolated from Tolypocladium sup_5 | | Descriptor: | PEPTAIBOL GICHIGAMIN | | Authors: | Du, L, Risinger, A.L, Mitchell, C.A, Stamps, B.W, Pan, N, King, J.B, Motley, J.L, Thomas, L.M, Yang, Z, Stevenson, B.S, Mooberry, S.L, Cichewicz, R.H. | | Deposit date: | 2015-03-26 | | Release date: | 2016-03-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peptaibol gichigamin isolated from Tolypocladium sup_5
TO BE PUBLISHED
|
|
4Z7A
 
 | | Structural and biochemical characterization of a non-functionally redundant M. tuberculosis (3,3) L,D-Transpeptidase, LdtMt5. | | Descriptor: | ACETYL GROUP, DI(HYDROXYETHYL)ETHER, Mycobacterium tuberculosis (3,3)L,D-Transpeptidase type 5, ... | | Authors: | Basta, L, Ghosh, A, Pan, Y, Jakoncic, J, Lloyd, E, Townsend, G, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-04-06 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Loss of a Functionally and Structurally Distinct ld-Transpeptidase, LdtMt5, Compromises Cell Wall Integrity in Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
4XBS
 
 | | 2-deoxyribose-5-phosphate aldolase mutant - E78K | | Descriptor: | Deoxyribose-phosphate aldolase | | Authors: | Jiao, X.-C, Pan, J, Xu, G.-C, Kong, X.-D, Chen, Q, Zhang, Z.-J, Xu, J.-H. | | Deposit date: | 2014-12-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Efficient synthesis of a statin precursor in high space-time yield by a new aldehyde-tolerant aldolase identified from Lactobacillus brevis
Catalysis Science And Technology, 5, 2015
|
|
4Z2D
 
 | | Quinolone(Levofloxacin)-DNA cleavage complex of gyrase from S. pneumoniae | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Selvarajah, J, Crevel, I.M.-T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2015-03-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structural studies of the drug-stabilized cleavage complexes of topoisomerase IV and gyrase from Streptococcus pneumoniae
To Be Published
|
|
4F62
 
 | | Crystal structure of a putative farnesyl-diphosphate synthase from Marinomonas sp. MED121 (Target EFI-501980) | | Descriptor: | CHLORIDE ION, GLYCEROL, Geranyltranstransferase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4Z2C
 
 | | Quinolone(Moxifloxacin)-DNA cleavage complex of gyrase from S. pneumoniae | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Selvarajah, J, Crevel, I.M.-T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2015-03-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural studies of the drug-stabilized cleavage complexes of topoisomerase IV and gyrase from Streptococcus pneumoniae
To Be Published
|
|
6DQ6
 
 | |
3RLJ
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with SARM S-22 | | Descriptor: | (2S)-3-(4-cyanophenoxy)-N-[4-cyano-3-(trifluoromethyl)phenyl]-2-hydroxy-2-methylpropanamide, Androgen receptor | | Authors: | Bohl, C.E, Duke, C.B, Jones, A, Dalton, J.T, Miller, D.D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unexpected binding orientation of bulky-B-ring anti-androgens and implications for future drug targets.
J.Med.Chem., 54, 2011
|
|
8TCD
 
 | | Structure of Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
5UGA
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium | | Descriptor: | 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
6QAD
 
 | | Human Butyrylcholinesterase in complex with ((S)-2-(butylamino)-N-(2-(4-(dimethylamino)cyclohexyl)ethyl)-3-(1H-indol-3-yl)propanamide | | Descriptor: | (2~{S})-2-(butylamino)-~{N}-[2-[4-(dimethylamino)cyclohexyl]ethyl]-3-(1~{H}-indol-3-yl)propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Nachon, F, Harst, M, Knez, D, Gobec, S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Tryptophan-derived butyrylcholinesterase inhibitors as promising leads against Alzheimer's disease.
Chem.Commun.(Camb.), 55, 2019
|
|
6Q54
 
 | | Structure of GluA2 ligand-binding domain (S1S2J) in complex with the agonist (S)-2-Amino-3-(1-ethyl-4-hydroxy-1H-1,2,3-triazol-5-yl)propanoic acid at 1.4 A resolution | | Descriptor: | (2~{S})-2-azanyl-3-(3-ethyl-5-oxidanyl-1,2,3-triazol-4-yl)propanoic acid, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Moellerud, S, Temperini, P, Kastrup, J.S. | | Deposit date: | 2018-12-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Use of the 4-Hydroxytriazole Moiety as a Bioisosteric Tool in the Development of Ionotropic Glutamate Receptor Ligands.
J.Med.Chem., 62, 2019
|
|
7UBO
 
 | | Crystal Structure of the first bromodomain of human BRDT in complex with the inhibitor CCD-956 | | Descriptor: | Bromodomain testis-specific protein, DIMETHYL SULFOXIDE, N-[(2R)-1-(methylamino)-3-{1-[(4-methyl-2-oxo-1,2-dihydroquinolin-6-yl)acetyl]piperidin-4-yl}-1-oxopropan-2-yl]-5-phenylpyridine-2-carboxamide | | Authors: | Ta, H.M, Modukuri, R.K, Yu, Z, Tan, Z, Matzuk, M.M, Kim, C. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of potent BET bromodomain 1 stereoselective inhibitors using DNA-encoded chemical library selections.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8T48
 
 | |
