1DJM
 
 | | SOLUTION STRUCTURE OF BEF3-ACTIVATED CHEY FROM ESCHERICHIA COLI | | Descriptor: | CHEMOTAXIS PROTEIN Y | | Authors: | Cho, H.S, Lee, S.Y, Yan, D, Pan, X, Parkinson, J.S, Kustu, S, Wemmer, D.E, Pelton, J.G. | | Deposit date: | 1999-12-03 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of activated CheY.
J.Mol.Biol., 297, 2000
|
|
4N6Y
 
 | | Pim1 Complexed with a phenylcarboxamide | | Descriptor: | 2-(acetylamino)-N-[2-(piperidin-1-yl)phenyl]-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
1CR9
 
 | | CRYSTAL STRUCTURE OF THE ANTI-PRION FAB 3F4 | | Descriptor: | FAB ANTIBODY HEAVY CHAIN, FAB ANTIBODY LIGHT CHAIN | | Authors: | Kanyo, Z.F, Pan, K.M, Williamson, R.A, Burton, D.R, Prusiner, S.B, Fletterick, R.J, Cohen, F.E. | | Deposit date: | 1999-08-14 | | Release date: | 2000-04-17 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibody binding defines a structure for an epitope that participates in the PrPC-->PrPSc conformational change.
J.Mol.Biol., 293, 1999
|
|
7NFF
 
 | | An octameric barrel state of a de novo coiled-coil assembly: CC-Type2-(LaId)4-I24A. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CC-Type2-(LaId)4-I24A, GLYCEROL | | Authors: | Rhys, G.G, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DL7
 
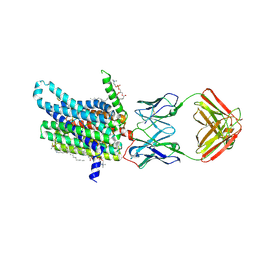 | | Cryo-EM structure of human ferroportin/slc40 bound to minihepcidin PR73 in nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 11F9 heavy-chain, 11F9 light-chain, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
8DL8
 
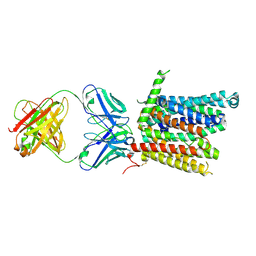 | | Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
8DL6
 
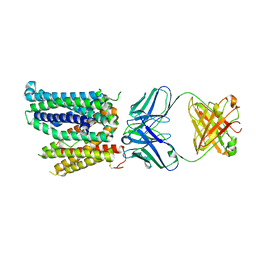 | | Cryo-EM structure of human ferroportin/slc40 bound to Ca2+ in nanodisc | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, CALCIUM ION, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of Ca 2+ transport by ferroportin.
Elife, 12, 2023
|
|
8RRZ
 
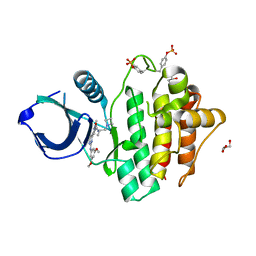 | | Crystal structure of SYK kinase in complex with compound 1 | | Descriptor: | GLYCEROL, N-[(2S)-1-(azetidin-1-yl)propan-2-yl]-3-{2-[(3,5-dimethoxyphenyl)amino]pyrimidin-4-yl}-1-methyl-1H-pyrazole-5-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Canevari, G. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and optimization of 4-pyrazolyl-2-aminopyrimidine derivatives as potent spleen tyrosine kinase (SYK) inhibitors.
Eur.J.Med.Chem., 270, 2024
|
|
7JWV
 
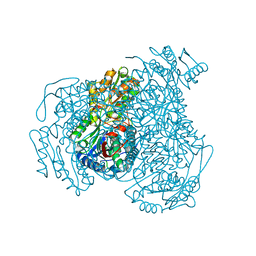 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 5-[4-(hydroxymethyl)phenyl]-1-methyl-6-{[(1R)-1-phenylethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7JWS
 
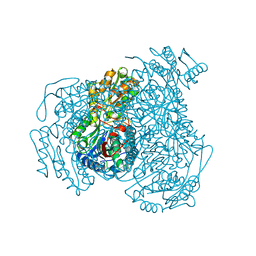 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 1-methyl-5-phenyl-6-{[(1R)-1-phenylethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7JWU
 
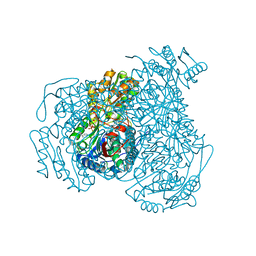 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 1-methyl-5-phenyl-6-{[(1R)-1-(pyridin-2-yl)ethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7JWT
 
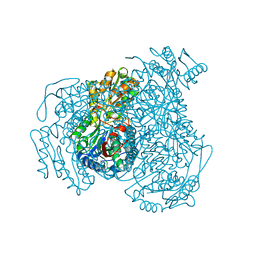 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 6-{[(1R)-1-(3-hydroxyphenyl)ethyl]sulfanyl}-1-methyl-5-phenyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7KEO
 
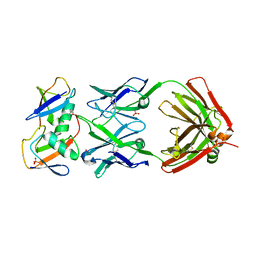 | | Crystal structure of K29-linked di-ubiquitin in complex with synthetic antigen binding fragment | | Descriptor: | PHOSPHATE ION, Synthetic antigen binding fragment, heavy chain, ... | | Authors: | Yu, Y, Zheng, Q, Erramilli, S, Pan, M, Kossiakoff, A, Liu, L, Zhao, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle.
Nat.Chem.Biol., 17, 2021
|
|
7C8D
 
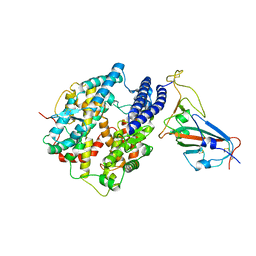 | | Cryo-EM structure of cat ACE2 and SARS-CoV-2 RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1, ZINC ION | | Authors: | Gao, G.F, Wang, Q.H, Wu, L.l. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2.
Cell Discov, 6, 2020
|
|
4Z80
 
 | |
4Z81
 
 | |
7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | Descriptor: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | Authors: | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
4UGQ
 
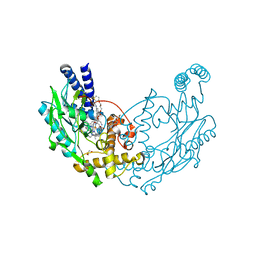 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with N,N''-(((2S)-3-aminopropane-1,2-diyl)bis(oxymethanediylbenzene-3,1- diyl))dithiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4ZSG
 
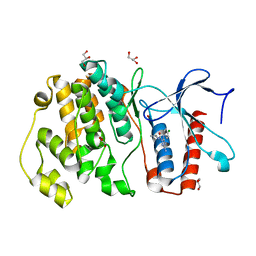 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | Descriptor: | 3-amino-5-[(4-chlorophenyl)amino]-N-(propan-2-yl)-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J, Ogg, D.J. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4ZSJ
 
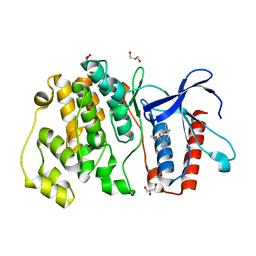 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | Descriptor: | 3-amino-5-[(4-chloro-3-methylphenyl)amino]-N-(propan-2-yl)-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J, Ogg, D.J. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4UGC
 
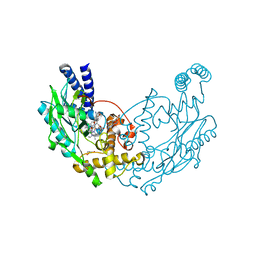 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6,6'-(((2S)-3-aminopropane-1,2-diyl)bis(oxymethanediyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-{[(2S)-3-aminopropane-1,2-diyl]bis(oxymethanediyl)}bis(4-methylpyridin-2-amine), CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
