2XXX
 
 | |
4U0D
 
 | | Hexameric HIV-1 CA in complex with Nup153 peptide, P212121 crystal form | | Descriptor: | CHLORIDE ION, Gag polyprotein, Nuclear pore complex protein Nup153 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
5OR9
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methyl-cyclopentapyrazole compound 13 | | Descriptor: | (2~{S})-1-(4-fluorophenyl)sulfonyl-~{N}-(2-methyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Vedove, A.D, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
4U0C
 
 | |
4U0B
 
 | |
4U0A
 
 | | Hexameric HIV-1 CA in complex with CPSF6 peptide, P6 crystal form | | Descriptor: | CHLORIDE ION, Capsid protein p24, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
1FCS
 
 | | CRYSTAL STRUCTURE OF A DISTAL SITE DOUBLE MUTANT OF SPERM WHALE MYOGLOBIN AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Rizzi, M, Bolognesi, M, Coda, A, Cutruzzola, F, Travaglini Allocatelli, C, Brancaccio, A, Brunori, M. | | Deposit date: | 1993-04-20 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a distal site double mutant of sperm whale myoglobin at 1.6 A resolution.
FEBS Lett., 320, 1993
|
|
2XXU
 
 | | Crystal structure of the GluK2 (GluR6) M770K LBD dimer in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
6HBT
 
 | |
5MVN
 
 | |
2XXT
 
 | | Crystal structure of the GluK2 (GluR6) wild-type LBD dimer in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, GLUTAMATE RECEPTOR, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
2XXV
 
 | | Crystal structure of the GluK2 (GluR6) M770K LBD dimer in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, GLUTAMATE RECEPTOR, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
5ORB
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methyl-cyclopentapyrazole compound 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-methoxyphenyl)sulfanyl-~{N}-(2-methyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)ethanamide, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Dalle Vedove, A, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
3QCE
 
 | |
4JFU
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor | | Descriptor: | (2S,3R,4S,5S)-2-methyl-5-(4-methylphenyl)pyrrolidine-3,4-diol, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
7F3Y
 
 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with methotrexate (MTX), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | Authors: | Vanichtanankul, J, Tanramluk, D, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
1XAI
 
 | | CRYSTAL STRUCTURE OF STAPHLYOCOCCUS AUREUS 3-DEHYDROQUINATE SYNTHASE (DHQS) IN COMPLEX WITH ZN2+, NAD+ AND CARBAPHOSPHONATE | | Descriptor: | 3-dehydroquinate synthase, ZINC ION, [1R-(1ALPHA,3BETA,4ALPHA,5BETA)]-5-(PHOSPHONOMETHYL)-1,3,4-TRIHYDROXYCYCLOHEXANE-1-CARBOXYLIC ACID | | Authors: | Nichols, C.E, Ren, J, Leslie, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-08-25 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparison of ligand induced conformational changes and domain closure mechanisms, between prokaryotic and eukaryotic dehydroquinate synthases.
J.Mol.Biol., 343, 2004
|
|
5OR8
 
 | | Crystal Structure of BAZ2A bromodomain in complex with 1,3-dimethyl-benzimidazolone compound 1 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[(4-fluorophenyl)methyl]-1,3,6-trimethyl-2-oxidanylidene-benzimidazole-5-sulfonamide | | Authors: | Lolli, G, Dalle Vedove, A, Marchand, J.-R, Caflisch, A. | | Deposit date: | 2017-08-15 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Inhibitors of Four Bromodomains by Fragment-Anchored Ligand Docking.
J Chem Inf Model, 57, 2017
|
|
5T2T
 
 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease bound to compound L742001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Alvarez, K, Ferron, F. | | Deposit date: | 2016-08-24 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.967 Å) | | Cite: | Crystal structures ofLymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
IUCrJ, 5, 2018
|
|
3MKS
 
 | | Crystal Structure of yeast Cdc4/Skp1 in complex with an allosteric inhibitor SCF-I2 | | Descriptor: | 1,1'-binaphthalene-2,2'-dicarboxylic acid, Cell division control protein 4, GLYCEROL, ... | | Authors: | Orlicky, S, Sicheri, F, Tyers, M, Tang, X. | | Deposit date: | 2010-04-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase.
Nat.Biotechnol., 28, 2010
|
|
2H5K
 
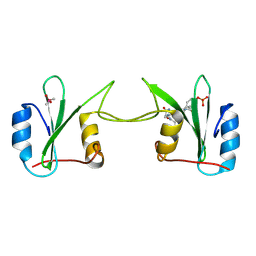 | | Crystal Structure of Complex Between the Domain-Swapped Dimeric Grb2 SH2 Domain and Shc-Derived Ligand, Ac-NH-pTyr-Val-Asn-NH2 | | Descriptor: | CACODYLATE ION, Growth factor receptor-bound protein 2, Shc-Derived Ligand | | Authors: | Benfield, A.P, Whiddon, B.B, Martin, S.F. | | Deposit date: | 2006-05-26 | | Release date: | 2006-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural and energetic aspects of Grb2-SH2 domain-swapping.
Arch.Biochem.Biophys., 462, 2007
|
|
5TDB
 
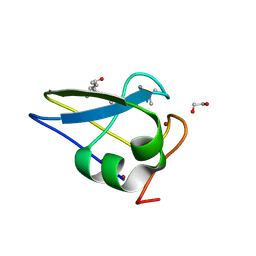 | | Crystal structure of the human UBR-box domain from UBR2 in complex with asymmetrically double methylated arginine peptide | | Descriptor: | 1,2-ETHANEDIOL, DA2-ILE-PHE-SER peptide, E3 ubiquitin-protein ligase UBR2, ... | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
7Z9L
 
 | | Phen-DC3 intercalation causes hybrid-to-antiparallel transformation of human telomeric DNA G-quadruplex | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N2,N9-bis(1-methylquinolin-3-yl)-1,10-phenanthroline-2,9-dicarboxamide | | Authors: | Ghosh, A, Trajkovski, M, Teulade-Fichou, M.P, Gabelica, V, Plavec, J. | | Deposit date: | 2022-03-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Phen-DC 3 Induces Refolding of Human Telomeric DNA into a Chair-Type Antiparallel G-Quadruplex through Ligand Intercalation.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1Y7D
 
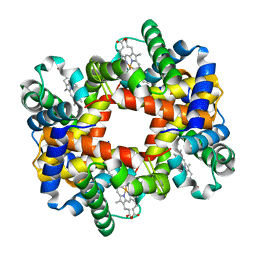 | |
1YE1
 
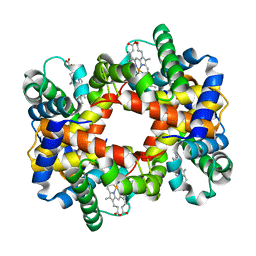 | | T-To-T(High) quaternary transitions in human hemoglobin: betaY35A oxy (2MM IHP, 20% PEG) (1 test set) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Rogers, P.H, Arnone, A. | | Deposit date: | 2004-12-27 | | Release date: | 2005-01-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Crystallographic evidence for a new ensemble of ligand-induced allosteric transitions in hemoglobin: the T-to-T(high) quaternary transitions.
Biochemistry, 44, 2005
|
|
