2PC2
 
 | | Lysozyme Cocrystallized with Tris-dipicolinate Eu complex | | Descriptor: | CHLORIDE ION, EUROPIUM ION, Lysozyme C, ... | | Authors: | Pompidor, G, Vicat, J, Kahn, R. | | Deposit date: | 2007-03-29 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | A dipicolinate lanthanide complex for solving protein structures using anomalous diffraction
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2OZ5
 
 | | Crystal structure of Mycobacterium tuberculosis protein tyrosine phosphatase PtpB in complex with the specific inhibitor OMTS | | Descriptor: | Phosphotyrosine protein phosphatase ptpb, {(3-CHLOROBENZYL)[(5-{[(3,3-DIPHENYLPROPYL)AMINO]SULFONYL}-2-THIENYL)METHYL]AMINO}(OXO)ACETIC ACID | | Authors: | Grundner, C, Gee, C.L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Selective Inhibition of Mycobacterium tuberculosis Protein Tyrosine Phosphatase PtpB.
Structure, 15, 2007
|
|
2PE7
 
 | | Thaumatin from Thaumatococcus Danielli in complex with tris-dipicolinate Europium | | Descriptor: | EUROPIUM ION, L(+)-TARTARIC ACID, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Pompidor, G, Vicat, J, Kahn, R. | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A dipicolinate lanthanide complex for solving protein structures using anomalous diffraction.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2PEK
 
 | | Crystal structure of RbcX point mutant Q29A | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
2PF0
 
 | |
2P2V
 
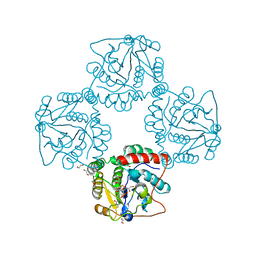 | | Crystal structure analysis of monofunctional alpha-2,3-sialyltransferase Cst-I from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, Alpha-2,3-sialyltransferase, CHLORIDE ION, ... | | Authors: | Chiu, C.P, Lairson, L.L, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2007-03-07 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of the alpha-2,3-Sialyltransferase Cst-I from Campylobacter jejuni in Apo and Substrate-Analogue Bound Forms.
Biochemistry, 46, 2007
|
|
1M01
 
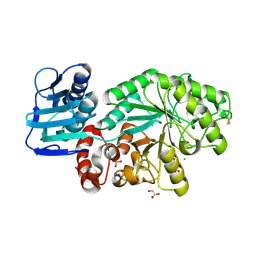 | | Wildtype Streptomyces plicatus beta-hexosaminidase in complex with product (GlcNAc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | J Williams, S, Mark, B.L, Vocadlo, D.J, James, M.N.G, Withers, S.G. | | Deposit date: | 2002-06-11 | | Release date: | 2003-01-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical
role in substrate-assisted catalysis by orienting the 2-acetamido group
and stabilizing the transition state.
J.Biol.Chem., 277, 2002
|
|
2P3H
 
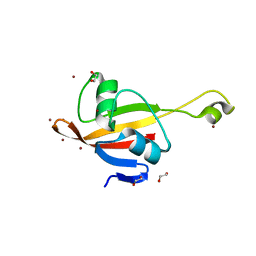 | | Crystal structure of the CorC_HlyC domain of a putative Corynebacterium glutamicum hemolysin | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Uncharacterized CBS domain-containing protein | | Authors: | Cuff, M.E, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the CorC_HlyC domain of a putative Corynebacterium glutamicum hemolysin.
TO BE PUBLISHED
|
|
2P59
 
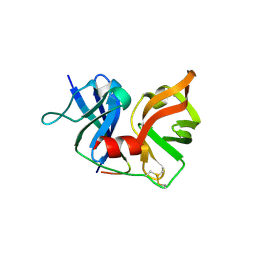 | | Crystal Structure of Hepatitis C Virus NS3.4A protease | | Descriptor: | (2S,3AS,7AS)-1-[(2S)-2-{[(2S)-2-CYCLOHEXYL-2-({[(2R)-4-NITRO-2H-PYRROL-2-YL]CARBONYL}AMINO)ACETYL]AMINO}-3,3-DIMETHYLBUTANOYL]-N-{(1S)-1-[(1R)-2-(CYCLOPROPYLAMINO)-1-HYDROXY-2-OXOETHYL]BUTYL}OCTAHYDRO-1H-INDOLE-2-CARBOXAMIDE, NS3, peptide | | Authors: | Perni, R.B, Wei, Y. | | Deposit date: | 2007-03-14 | | Release date: | 2008-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibitors of hepatitis C virus NS3.4A protease. Effect of P4 capping groups on inhibitory potency and pharmacokinetics.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2P5M
 
 | | C-terminal domain hexamer of AhrC bound with L-arginine | | Descriptor: | ARGININE, Arginine repressor | | Authors: | Garnett, J.A, Baumberg, S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2007-03-15 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the C-terminal effector-binding domain of AhrC bound to its corepressor L-arginine.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2P5Y
 
 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
2P6A
 
 | | The structure of the Activin:Follistatin 315 complex | | Descriptor: | Follistatin, Inhibin beta A chain, probable fragment of follistatin | | Authors: | Lerch, T.F, Shimasaki, S, Woodruff, T.K, Jardetzky, T.S. | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and biophysical coupling of heparin and activin binding to follistatin isoform functions.
J.Biol.Chem., 282, 2007
|
|
2P6R
 
 | |
2P5W
 
 | | Crystal structures of high affinity human T-cell receptors bound to pMHC reveal native diagonal binding geometry | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, CALCIUM ION, ... | | Authors: | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | Deposit date: | 2007-03-16 | | Release date: | 2007-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
8PWH
 
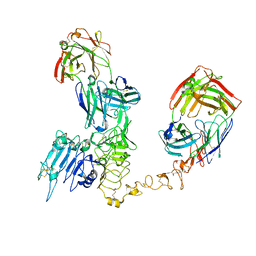 | | Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, ... | | Authors: | Ruedas, R, Vuillemot, R, Tubiana, T, Winter, J.M, Pieri, L, Arteni, A.A, Samson, C, Jonic, J, Mathieu, M, Bressanelli, S. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex.
J.Struct.Biol., 216, 2024
|
|
2P6Z
 
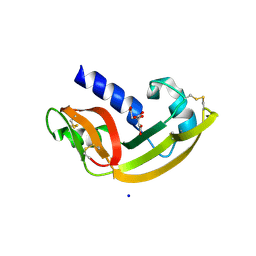 | |
2P83
 
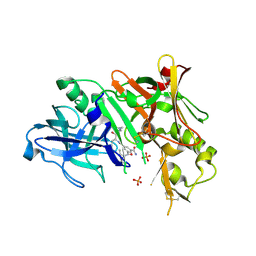 | | Potent and selective isophthalamide S2 hydroxyethylamine inhibitor of BACE1 | | Descriptor: | Beta-secretase 1, N~3~-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-N~1~,N~1~-DIPROPYLBENZENE-1,3,5-TRICARBOXAMIDE, PHOSPHATE ION | | Authors: | Benson, T.E, Prince, D.B, Tomasselli, A.G, Emmons, T.L, Paddock, D.J. | | Deposit date: | 2007-03-21 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Potent and selective isophthalamide S(2) hydroxyethylamine inhibitors of BACE1.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2P8V
 
 | | Crystal structure of human Homer3 EVH1 domain | | Descriptor: | Homer protein homolog 3, SULFATE ION | | Authors: | Bouyain, S, Tu, J, Huang, G.N, Worley, P.F, Leahy, D. | | Deposit date: | 2007-03-23 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | NFAT binding and regulation of T cell activation by the cytoplasmic scaffolding Homer proteins.
Science, 319, 2008
|
|
2P93
 
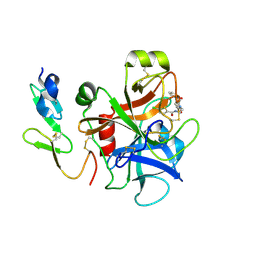 | |
2P8R
 
 | | Crystal structure of the C-terminal domain of C. elegans pre-mRNA splicing factor Prp8 carrying R2303K mutant | | Descriptor: | Pre-mRNA-splicing factor Prp8 | | Authors: | Zhang, L, Shen, J, Guarnieri, M.T, Heroux, A, Yang, K, Zhao, R. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
Protein Sci., 16, 2007
|
|
2P94
 
 | |
2P9T
 
 | | Crystal Structure of Phosphoglycerate Kinase-2 bound to 3-phosphoglycerate | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Phosphoglycerate kinase, testis specific | | Authors: | Sawyer, G.M, Monzingo, A.F, Poteet, E.C, Robertus, J.D. | | Deposit date: | 2007-03-26 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analysis of phosphoglycerate kinase 2, a sperm-specific isoform from Mus musculus.
Proteins, 71, 2007
|
|
8ID7
 
 | |
2P9A
 
 | | E. coli methionine aminopeptidase dimetalated with inhibitor YE6 | | Descriptor: | 5-(2-chlorophenyl)furan-2-carbohydrazide, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q. | | Deposit date: | 2007-03-24 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of Monometalated Methionine Aminopeptidase: Inhibitor Discovery and Crystallographic Analysis.
J.Med.Chem., 50, 2007
|
|
2PBI
 
 | | The multifunctional nature of Gbeta5/RGS9 revealed from its crystal structure | | Descriptor: | GLYCEROL, Guanine nucleotide-binding protein subunit beta 5, Regulator of G-protein signaling 9 | | Authors: | Cheever, M.L, Snyder, J.T, Gershburg, S, Siderovski, D.P, Harden, T.K, Sondek, J. | | Deposit date: | 2007-03-28 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the multifunctional Gbeta5-RGS9 complex.
Nat.Struct.Mol.Biol., 15, 2008
|
|
