8X2A
 
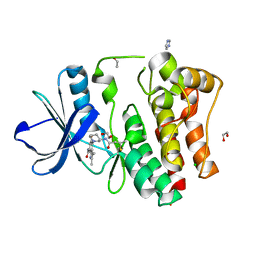 | | The Crystal Structure of BMX from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3S)-3-{[(2E)-but-2-enoyl]amino}piperidin-1-yl]-5-fluoro-2,3-dimethyl-1H-indole-7-carboxamide, CHLORIDE ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2023-11-09 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Crystal Structure of BMX from Biortus.
To Be Published
|
|
2GNI
 
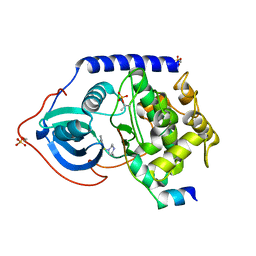 | | PKA fivefold mutant model of Rho-kinase with inhibitor Fasudil (HA1077) | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, cAMP-dependent protein kinase inhibitor alpha, cAMP-dependent protein kinase, ... | | Authors: | Bonn, S, Herrero, S, Breitenlechner, C.B, Engh, R.A, Gassel, M, Bossemeyer, D. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural analysis of protein kinase A mutants with Rho-kinase inhibitor specificity
J.Biol.Chem., 281, 2006
|
|
8V4X
 
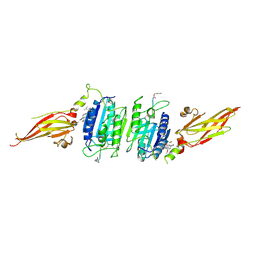 | | Structure of MALT1 in complex with an allosteric inhibitor | | Descriptor: | Inhibitor peptide, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, N-{7-[(1S)-1-methoxyethyl]-2-methyl[1,3]thiazolo[5,4-b]pyridin-6-yl}-N'-[6-(2H-1,2,3-triazol-2-yl)-5-(trifluoromethyl)pyridin-3-yl]urea | | Authors: | Judge, R.A, Pappano, W.N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Inhibition of MALT1 and BCL2 Induces Synergistic Antitumor Activity in Models of B-Cell Lymphoma.
Mol.Cancer Ther., 23, 2024
|
|
8YR2
 
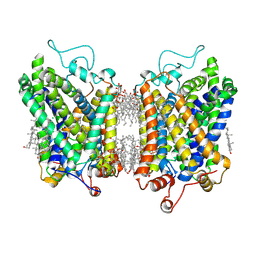 | | Structure of NET-Nisoxetine in outward-open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y93
 
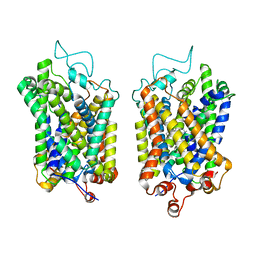 | | Structure of NET-Amitriptyline in outward-open state | | Descriptor: | Amitriptyline, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8TR3
 
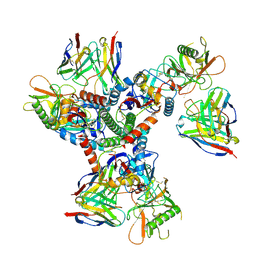 | | Cryo-EM structure of HmAb64 scFv in complex with CNE40 SOSIP trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CNE40 SOSIP Envelope glycoprotein gp120, ... | | Authors: | Chan, K.-W, Kong, X.P. | | Deposit date: | 2023-08-09 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Human CD4-binding site antibody elicited by polyvalent DNA prime-protein boost vaccine neutralizes cross-clade tier-2-HIV strains.
Nat Commun, 15, 2024
|
|
5HZC
 
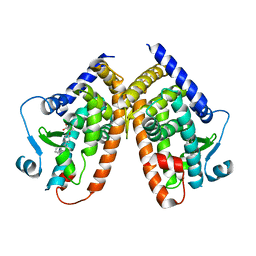 | | Crystal structure of the complex PPARgamma/AL26-29 | | Descriptor: | 2-methyl-2-[4-(naphthalen-1-yl)phenoxy]propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Lavecchia, A. | | Deposit date: | 2016-02-02 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for PPAR partial or full activation revealed by a novel ligand binding mode.
Sci Rep, 6, 2016
|
|
5HYK
 
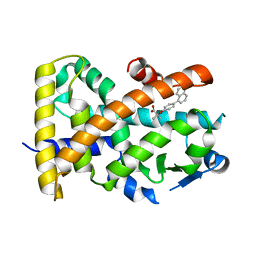 | | Crystal structure of the complex PPARalpha/AL26-29 | | Descriptor: | 2-methyl-2-[4-(naphthalen-1-yl)phenoxy]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Lavecchia, A. | | Deposit date: | 2016-02-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for PPAR partial or full activation revealed by a novel ligand binding mode.
Sci Rep, 6, 2016
|
|
7NA1
 
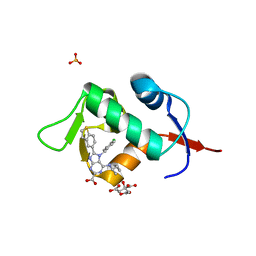 | | HDM2 in complex with compound 2 | | Descriptor: | 8-(1-benzothiophen-5-yl)-7-[(4-chlorophenyl)methyl]-6-{[(1R)-1-cyclopropylethyl]amino}-7H-purine-2-carboxylic acid, CITRIC ACID, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7NA4
 
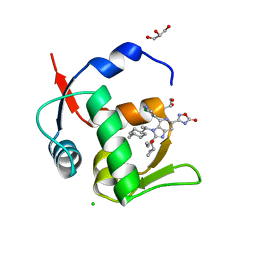 | | HDM2 in complex with compound 63 | | Descriptor: | 3-[4-(5-chloropyridin-3-yl)-2-[(R)-cyclopropyl(ethoxy)methyl]-3-{(1R)-1-[(1r,4R)-4-methylcyclohexyl]ethyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
6DB7
 
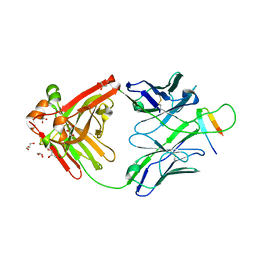 | |
6O07
 
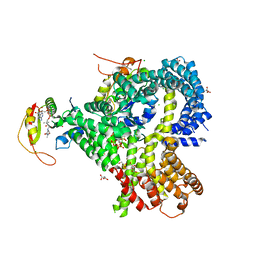 | |
4QU1
 
 | |
8HUQ
 
 | | X-ray structure of human PPAR alpha ligand binding domain-elafibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[2,6-dimethyl-4-[(~{E})-3-(4-methylsulfanylphenyl)-3-oxidanylidene-prop-1-enyl]phenoxy]-2-methyl-propanoic acid, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
8IFQ
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 1 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGY
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
6ASQ
 
 | | Structure of Grp94 bound to methyl 2-[2-(2-benzylpyridin-3-yl)ethyl]-3-chloro-4,6-dihydroxybenzoate, a pan-Hsp90 inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, Endoplasmin, ... | | Authors: | Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2017-08-25 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Trifunctional High-Throughput Screen Identifies Promising Scaffold To Inhibit Grp94 and Treat Myocilin-Associated Glaucoma.
ACS Chem. Biol., 13, 2018
|
|
3TLQ
 
 | | Crystal structure of EAL-like domain protein YdiV | | Descriptor: | GLYCEROL, PHOSPHATE ION, Regulatory protein YdiV | | Authors: | Li, B, Li, N, Wang, F, Guo, L, Liu, C, Zhu, D, Xu, S, Gu, L. | | Deposit date: | 2011-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
3QQV
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from corynebacterium glutamicum complexed with isoprenyl diphosphate and magnesium | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, Geranylgeranyl pyrophosphate synthase, ... | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Poulter, C.D, Gerlt, J.A, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-02-16 | | Release date: | 2011-03-02 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3RO2
 
 | | Structures of the LGN/NuMA complex | | Descriptor: | G-protein-signaling modulator 2, GLYCEROL, peptide of Nuclear mitotic apparatus protein 1 | | Authors: | Shang, Y, Wei, Z. | | Deposit date: | 2011-04-25 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and G[alpha]i/LGN/NuMA pathways
Mol.Cell, 43, 2011
|
|
3P41
 
 | | Crystal structure of polyprenyl synthetase from pseudomonas fluorescens pf-5 complexed with magnesium and isoprenyl pyrophosphate | | Descriptor: | CHLORIDE ION, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-05 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3PDE
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from Lactobacillus brevis atcc 367 complexed with isoprenyl diphosphate and magnesium | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Farnesyl-diphosphate synthase, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3Q2Q
 
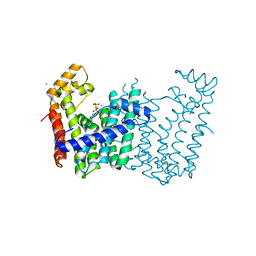 | | Crystal structure of geranylgeranyl pyrophosphate synthase from Corynebacterium glutamicum complexed with calcium and isoprenyl diphosphate | | Descriptor: | CALCIUM ION, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Poulter, C.D, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-12-20 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
