4DMI
 
 | | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences (CASP) | | Descriptor: | 1,2-ETHANEDIOL, Capsid Protein, SODIUM ION | | Authors: | Craig, T.K, Abendroth, J, Lorimer, D, Burgin Jr, A.B, Segall, A, Rohwer, F. | | Deposit date: | 2012-02-07 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences
To be Published
|
|
4MB4
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MCA
 
 | |
4MCH
 
 | | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with 6-hydroxy-1-naphthoic acid, NYSGRC Target 029520. | | Descriptor: | 6-hydroxynaphthalene-1-carboxylic acid, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with 6-hydroxy-1-naphthoic acid, NYSGRC Target 029520.
To be Published
|
|
4DGM
 
 | | Crystal Structure of maize CK2 in complex with the inhibitor apigenin | | Descriptor: | 1,2-ETHANEDIOL, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Casein kinase II subunit alpha, ... | | Authors: | Lolli, G, Mazzorana, M, Battistutta, R. | | Deposit date: | 2012-01-26 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of protein kinase CK2 by flavonoids and tyrphostins. A structural insight.
Biochemistry, 51, 2012
|
|
4DHK
 
 | |
4MEI
 
 | |
4DI0
 
 | |
4MDM
 
 | | Nido-Carborane Carbonic Anhydrase Inhibitor | | Descriptor: | 7-(sulfamoylamino)methyl-7,8-dicarba-nido-undecaborane, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Mader, P, Rezacova, P, Cigler, P, Gruner, B. | | Deposit date: | 2013-08-23 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carborane-based carbonic anhydrase inhibitors.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4DIE
 
 | |
4MEP
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3-chloro-pyridone ligand | | Descriptor: | 3-chloro-5-[1-(3-methylpyridin-2-yl)-3-phenyl-1H-1,2,4-triazol-5-yl]pyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
4DQM
 
 | | Revealing a marine natural product as a novel agonist for retinoic acid receptors with a unique binding mode and antitumor activity | | Descriptor: | (5S)-4-[(3E,7E)-4,8-dimethyl-10-(2,6,6-trimethylcyclohex-1-en-1-yl)deca-3,7-dien-1-yl]-5-hydroxyfuran-2(5H)-one, Nuclear receptor coactivator 1, Retinoic acid receptor alpha | | Authors: | Wang, S, Wang, Z, Lin, S, Zheng, W, Wang, R, Jin, S, Chen, J, Jin, L, Li, Y. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Revealing a natural marine product as a novel agonist for retinoic acid receptors with a unique binding mode and inhibitory effects on cancer cells.
Biochem.J., 446, 2012
|
|
4MDX
 
 | |
4DK9
 
 | | Crystal Structure of MBD4 Catalytic Domain Bound to Abasic DNA | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*T)-3', Methyl-CpG-binding domain protein 4, ... | | Authors: | Manvilla, B.A, Toth, E.A, Drohat, A.C. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal Structure of Human Methyl-Binding Domain IV Glycosylase Bound to Abasic DNA.
J.Mol.Biol., 420, 2012
|
|
4MEO
 
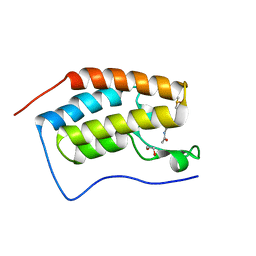 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-methyl-quinoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
4DLI
 
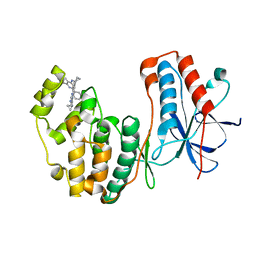 | | Human p38 MAP kinase in complex with RL87 | | Descriptor: | Mitogen-activated protein kinase 14, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | Authors: | Gruetter, C, Getlik, M, Simard, J.R, Rauh, D. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Fluorophore labeled kinase detects ligands that bind within the MAPK insert of p38alpha kinase.
Plos One, 7, 2012
|
|
4MFV
 
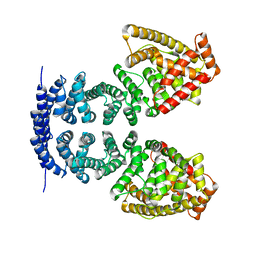 | | Crystal structure of human CTNNBL1(residues 33~563) | | Descriptor: | Beta-catenin-like protein 1 | | Authors: | Ahn, J.W, Kim, S, Kim, K.J. | | Deposit date: | 2013-08-28 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural insights into the novel ARM-repeat protein CTNNBL1 and its association with the hPrp19-CDC5L complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4DAR
 
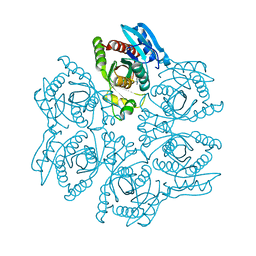 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Purine nucleoside phosphorylase deoD-type | | Authors: | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4LP0
 
 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid, Topoisomerase IV subunit B | | Authors: | Basarab, G.S, Manchester, J.I, Bist, S, Boriack-Sjodin, P.A, Dangel, B, Illingsworth, R, Uria-Nickelsen, M, Sherer, B.A, Sriram, S, Eakin, A.E. | | Deposit date: | 2013-07-14 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents.
J.Med.Chem., 56, 2013
|
|
4DNI
 
 | | Structure of Editosome protein | | Descriptor: | Fusion protein of RNA-editing complex proteins MP42 and MP18 | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-08 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Explorations of linked editosome domains leading to the discovery of motifs defining conserved pockets in editosome OB-folds.
J.Struct.Biol., 180, 2012
|
|
4N3Y
 
 | |
4N4H
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
4MZA
 
 | | Crystal structure of hPIV3 hemagglutinin-neuraminidase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2013-09-29 | | Release date: | 2013-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Interaction between the hemagglutinin-neuraminidase and fusion glycoproteins of human parainfluenza virus type III regulates viral growth in vivo.
MBio, 4, 2013
|
|
4N9T
 
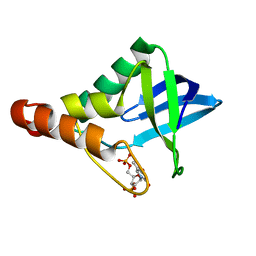 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66A/I92S at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-10-21 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cavities in proteins
To be Published
|
|
4N43
 
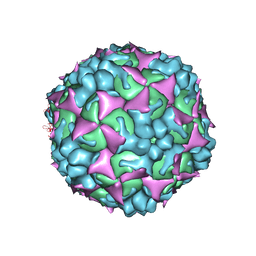 | |
