3F2C
 
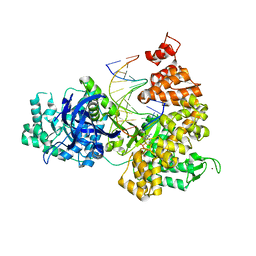 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP and Mn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3F2D
 
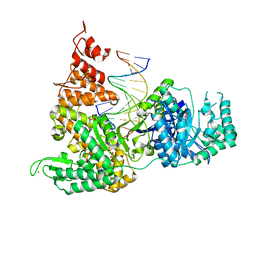 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP, Mn and Zn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4IL1
 
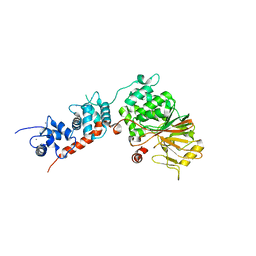 | | Crystal Structure of the Rat Calcineurin | | Descriptor: | CALCIUM ION, Calmodulin, Calcineurin subunit B type 1, ... | | Authors: | Ye, Q, Faucher, F, Jia, Z. | | Deposit date: | 2012-12-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of calcineurin activation by calmodulin.
Cell Signal, 25, 2013
|
|
7TM3
 
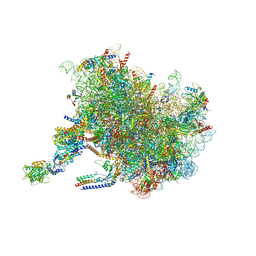 | | Structure of the rabbit 80S ribosome stalled on a 2-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
7TUT
 
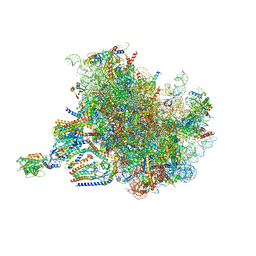 | | Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-02-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
6CP0
 
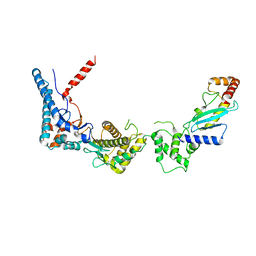 | | SdcA in complex with the E2, UbcH5C | | Descriptor: | SdcA, Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Wasilko, D.J, Huang, Q, Mao, Y. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Insights into the ubiquitin transfer cascade catalyzed by theLegionellaeffector SidC.
Elife, 7, 2018
|
|
1JVP
 
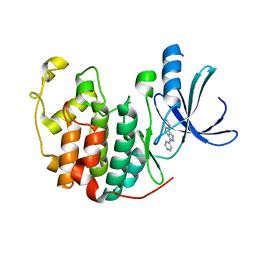 | |
7WTW
 
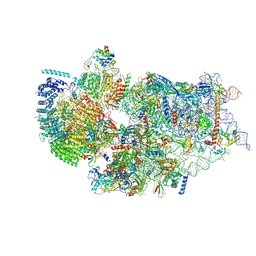 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A3 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
8Q7N
 
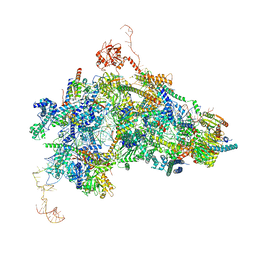 | | cryo-EM structure of the human spliceosomal B complex protomer (tri-snRNP core region) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, MINX pre-mRNA, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2023-08-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | New insights into the functions of B complex proteins revealed by cryo-EM of dimerized spliceosomes
To Be Published
|
|
6W0L
 
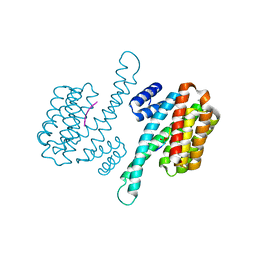 | | Henipavirus W protein interacts with 14-3-3 to modulate host gene expression | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, Phosphorylated W peptide | | Authors: | Edwards, M, Hoad, M, Tsimbalyuk, S, Menicucci, A, Messaoudi, I, Forwood, J, Basler, C. | | Deposit date: | 2020-03-01 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Henipavirus W Proteins Interact with 14-3-3 To Modulate Host Gene Expression.
J.Virol., 94, 2020
|
|
7QWS
 
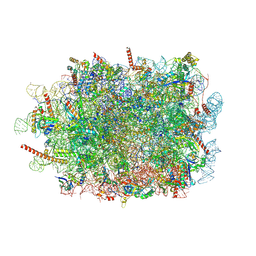 | | Structure of ribosome translating beta-tubulin in complex with TTC5 and NAC | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
1K2H
 
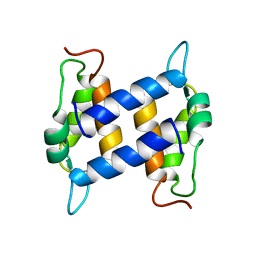 | | Three-dimensional Solution Structure of apo-S100A1. | | Descriptor: | S-100 protein, alpha chain | | Authors: | Rustandi, R.R, Baldisseri, D.M, Inman, K.G, Nizner, P, Hamilton, S.M, Landar, A, Landar, A, Zimmer, D.B, Weber, D.J. | | Deposit date: | 2001-09-27 | | Release date: | 2002-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium-signaling protein apo-S100A1 as determined by NMR.
Biochemistry, 41, 2002
|
|
7WTT
 
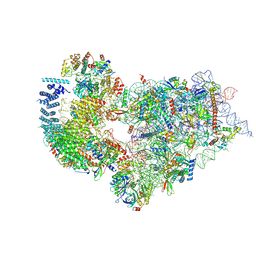 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A1 (with CK1) | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTU
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A1 (without CK1) | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
4IB0
 
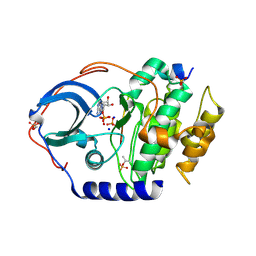 | |
4IAZ
 
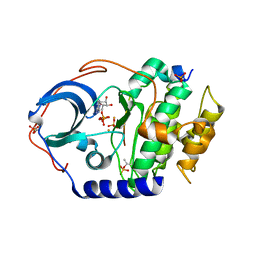 | |
5CYI
 
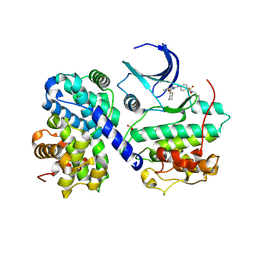 | | CDK2/Cyclin A covalent complex with 6-(cyclohexylmethoxy)-N-(4-(vinylsulfonyl)phenyl)-9H-purin-2-amine (NU6300) | | Descriptor: | 6-(cyclohexylmethoxy)-N-[4-(ethylsulfonyl)phenyl]-9H-purin-2-amine, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Anscombe, E, Meschini, E, Vidal, R.M, Martin, M.P, Staunton, D, Geitmann, M, Danielson, U.H, Stanley, W.A, Wang, L.Z, Reuillon, T, Golding, B.T, Cano, C, Newell, D.R, Noble, M.E.M, Wedge, S.R, Endicott, J.A, Griffin, R.J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Characterization of an Irreversible Inhibitor of CDK2.
Chem.Biol., 22, 2015
|
|
6IRL
 
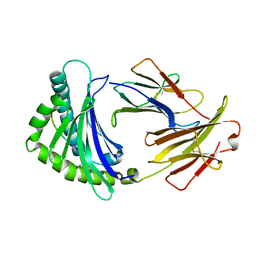 | |
1K6X
 
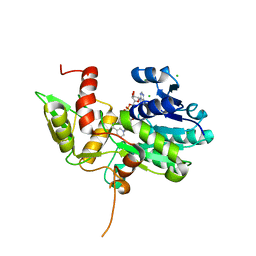 | | Crystal structure of Nmra, a negative transcriptional regulator in complex with NAD at 1.5 A resolution (Trigonal form) | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2002
|
|
7WTV
 
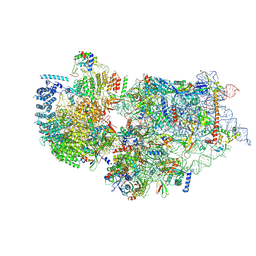 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A2 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7OYD
 
 | | Cryo-EM structure of a rabbit 80S ribosome with zebrafish Dap1b | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
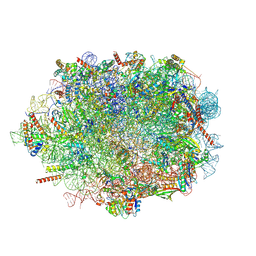
7OW7
 
 | |
7TOQ
 
 | | Mammalian 80S ribosome bound with the ALS/FTD-associated dipeptide repeat protein poly-PR | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
7AYF
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound TCF521-110 | | Descriptor: | 14-3-3 protein sigma, 6-(2-bromanylimidazol-1-yl)pyridine-3-carbaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7AZ2
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1014 | | Descriptor: | 1-[4-methyl-2-(trifluoromethyl)phenyl]-2-phenyl-imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
