4NHV
 
 | |
3O6E
 
 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4NIL
 
 | |
4JLP
 
 | |
1XON
 
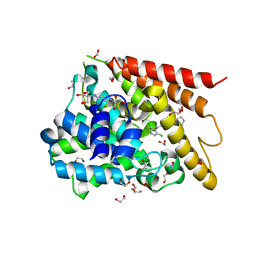 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With Piclamilast | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(CYCLOPENTYLOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-METHOXYBENZAMIDE, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-06 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
6EWE
 
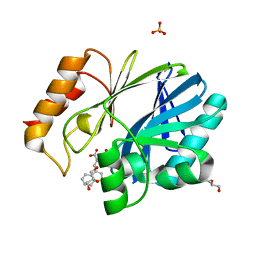 | | Crystal structure of BCII Metallo-beta-lactamase in complex with DZ-308 | | Descriptor: | (~{Z})-3-(1-benzothiophen-3-yl)-2-sulfanyl-prop-2-enoic acid, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Stepanovs, D, McDonough, M.A, Schofield, C.J, Zhang, D, El-Husseiny, A, Brem, J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure activity relationship studies on rhodanines and derived enethiol inhibitors of metallo-beta-lactamases.
Bioorg. Med. Chem., 26, 2018
|
|
2VHC
 
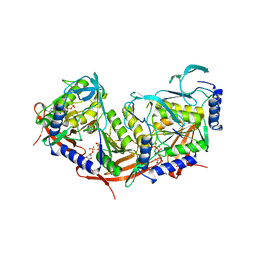 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 N234G mutant in complex with AMPCPP and MN | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
3OD5
 
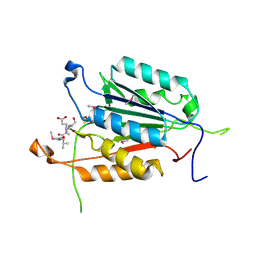 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO | | Descriptor: | CACODYLATE ION, Caspase-6, peptide aldehyde inhibitor AC-VEID-CHO | | Authors: | Wang, X.-J, Liu, X, Wang, K.-T, Cao, Q, Su, X.-D. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
2XYY
 
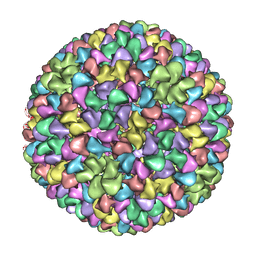 | | De Novo model of Bacteriophage P22 procapsid coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2NUI
 
 | | X-ray Structure of synthetic [D83A]RNase A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Boerema, D.J, Tereshko, V.A, Kent, S.B.H. | | Deposit date: | 2006-11-09 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Total synthesis by modern chemical ligation methods and high resolution (1.1 A) X-ray structure of ribonuclease A
Biopolymers, 90, 2008
|
|
2OJP
 
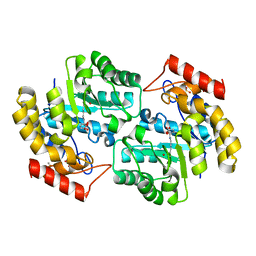 | | The crystal structure of a dimeric mutant of Dihydrodipicolinate synthase from E.coli- DHDPS-L197Y | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL | | Authors: | Griffin, M.D.W, Dobson, R.C.J, Antonio, L, Perugini, M.A, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2007-01-13 | | Release date: | 2008-01-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of quaternary structure in a homotetrameric enzyme.
J.Mol.Biol., 380, 2008
|
|
2OK4
 
 | |
6F13
 
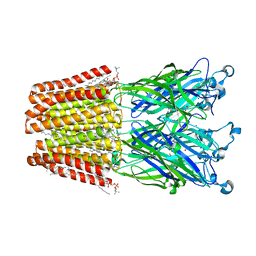 | | GLIC mutant E75A | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
9D77
 
 | | Crystal form of Netrin-1 mimics nanotubes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Meier, M, Krahn, N.J, McDougall, M.D, Rafiei, F, Koch, M, Stetefeld, J. | | Deposit date: | 2024-08-16 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanistic insights in the higher-order protein assemblies of netrin-1
To Be Published
|
|
3OEV
 
 | | Structure of yeast 20S open-gate proteasome with Compound 25 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(benzyloxy)-N-[(2S,3R)-3-hydroxy-1-{[(2S)-1-{[(3-methylthiophen-2-yl)methyl]amino}-1-oxo-4-phenylbutan-2-yl]amino}-1-oxobutan-2-yl]benzamide, MAGNESIUM ION, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-08-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5PZS
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[6-[(3-chlorophenyl)sulfonylcarbamoylamino]hexyl]urea | | Descriptor: | Fructose-1,6-bisphosphatase 1, N,N'-(hexane-1,6-diyldicarbamoyl)bis(3-chlorobenzene-1-sulfonamide) | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-11 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(3-chlorophenyl)sulfonyl-3-[6-[(3-chlorophenyl)sulfonylcarbamoylamino]hexyl]urea
To be published
|
|
1TY2
 
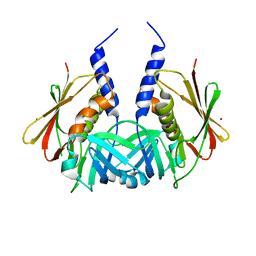 | | Crystal structure of the streptococcal pyrogenic exotoxin J (SPE-J) | | Descriptor: | ZINC ION, putative exotoxin (superantigen) | | Authors: | Baker, H.M, Proft, T, Webb, P.D, Arcus, V.L, Fraser, J.D, Baker, E.N. | | Deposit date: | 2004-07-07 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and mutational data show that the streptococcal pyrogenic exotoxin j can use a common binding surface for T-cell receptor binding and dimerization
J.Biol.Chem., 279, 2004
|
|
1BUE
 
 | | NMC-A CARBAPENEMASE FROM ENTEROBACTER CLOACAE | | Descriptor: | PROTEIN (IMIPENEM-HYDROLYSING BETA-LACTAMASE) | | Authors: | Swaren, P, Maveyraud, L, Cabantous, S, Pedelacq, J.D, Mourey, L, Frere, J.M, Samama, J.P. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray analysis of the NMC-A beta-lactamase at 1.64-A resolution, a class A carbapenemase with broad substrate specificity.
J.Biol.Chem., 273, 1998
|
|
2NYZ
 
 | |
2I0T
 
 | |
1BUZ
 
 | | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SPOIIAA | | Authors: | Kovacs, H, Comfort, D, Lord, M, Campbell, I.D, Yudkin, M.D. | | Deposit date: | 1997-09-08 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1L2J
 
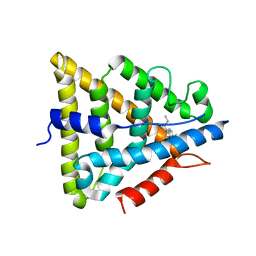 | | Human Estrogen Receptor beta Ligand-binding Domain in Complex with (R,R)-5,11-cis-diethyl-5,6,11,12-tetrahydrochrysene-2,8-diol | | Descriptor: | (R,R)-5,11-CIS-DIETHYL-5,6,11,12-TETRAHYDROCHRYSENE-2,8-DIOL, ESTROGEN RECEPTOR BETA | | Authors: | Shiau, A.K, Barstad, D, Radek, J.T, Meyers, M.J, Nettles, K.W, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Agard, D.A, Greene, G.L. | | Deposit date: | 2002-02-21 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism.
Nat.Struct.Biol., 9, 2002
|
|
4NUT
 
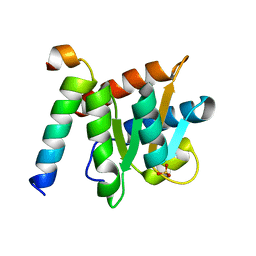 | | Crystal structure of the complex between Snu13p and the PEP domain of Rsa1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, Ribosome assembly 1 protein, SULFATE ION | | Authors: | Charron, C, Chagot, M.E, Manival, X, Branlant, C, Charpentier, B. | | Deposit date: | 2013-12-04 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control
J.Cell Biol., 207, 2014
|
|
3V6Z
 
 | | Crystal Structure of Hepatitis B Virus e-antigen | | Descriptor: | Fab e6 Heavy Chain, Fab e6 Light Chain, e-antigen | | Authors: | Dimattia, M.A, Watts, N.R, Stahl, S.J, Grimes, J.M, Steven, A.C, Stuart, D.I, Wingfield, P.T. | | Deposit date: | 2011-12-20 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein.
Structure, 21, 2013
|
|
3OC3
 
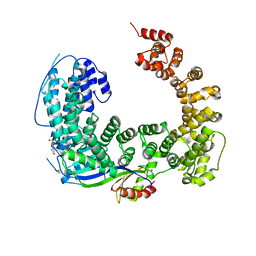 | | Crystal structure of the Mot1 N-terminal domain in complex with TBP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HELICASE MOT1, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Wollmann, P, Cui, S, Viswanathan, R, Berninghausen, O, Wells, M.N, Moldt, M, Witte, G, Butryn, A, Wendler, P, Beckmann, R, Auble, D.T, Hopfner, K.-P. | | Deposit date: | 2010-08-09 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
