4C4W
 
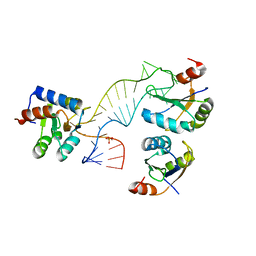 | | Structure of a rare, non-standard sequence k-turn bound by L7Ae protein | | Descriptor: | 50S RIBOSOMAL PROTEIN L7AE, DIHYDROGENPHOSPHATE ION, TSKT-23, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2013-09-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a Rare, Non-Standard Sequence K-Turn Bound by L7Ae Protein
Nucleic Acids Res., 42, 2014
|
|
6NCQ
 
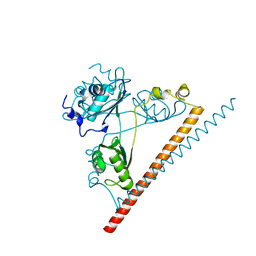 | | The dimerization domain of human SFPQ in space group C2221 | | Descriptor: | Splicing factor, proline- and glutamine-rich | | Authors: | Lee, M. | | Deposit date: | 2018-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new crystal structure and small-angle X-ray scattering analysis of the homodimer of human SFPQ.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6OWJ
 
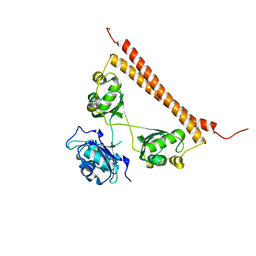 | | Zn-mediated polymerization of human SFPQ | | Descriptor: | Splicing factor, proline- and glutamine-rich, ZINC ION | | Authors: | Lee, M. | | Deposit date: | 2019-05-10 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis of the zinc-induced cytoplasmic aggregation of the RNA-binding protein SFPQ.
Nucleic Acids Res., 48, 2020
|
|
5IFM
 
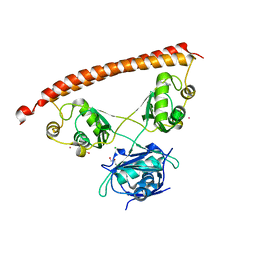 | | Human NONO (p54nrb) Homodimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-POU domain-containing octamer-binding protein, ... | | Authors: | Knott, G.J, Bond, C.S. | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic study of human NONO (p54(nrb)): overcoming pathological problems with purification, data collection and noncrystallographic symmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5IFN
 
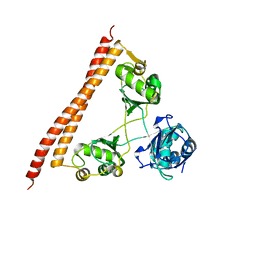 | |
4A8X
 
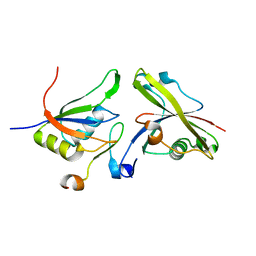 | | Structure of the core ASAP complex | | Descriptor: | HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18, HOOK-LIKE, ISOFORM A, ... | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
4CQ1
 
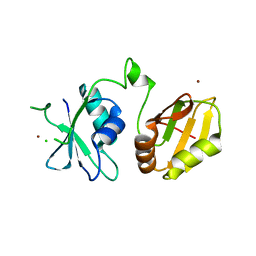 | | Crystal structure of the neuronal isoform of PTB | | Descriptor: | CHLORIDE ION, POLYPYRIMIDINE TRACT-BINDING PROTEIN 2, ZINC ION | | Authors: | Joshi, A, Buckroyd, A.N, Curry, S. | | Deposit date: | 2014-02-10 | | Release date: | 2014-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Solution and Crystal Structures of a C Terminal Fragment of the Neuronal Isoform of the Polypyrimidine Tract Binding Protein (Nptb)
Peerj, 2, 2014
|
|
8ANY
 
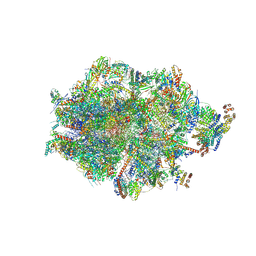 | | Human mitochondrial ribosome in complex with LRPPRC, SLIRP, A-site, P-site, E-site tRNAs and mRNA | | Descriptor: | 1,4-DIAMINOBUTANE, 12S mitochondrial rRNA, 16S mitochondrial rRNA, ... | | Authors: | Singh, V, Itoh, Y, Amunts, A. | | Deposit date: | 2022-08-06 | | Release date: | 2023-08-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of LRPPRC-SLIRP-dependent translation by the
mitoribosome
To Be Published
|
|
8BY6
 
 | | Structure of the human nuclear cap-binding complex bound to NCBP3(560-620) and cap-analogue m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2022-12-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
8CG3
 
 | |
8CGG
 
 | |
8CGH
 
 | |
5A5U
 
 | | Structure of mammalian eIF3 in the context of the 43S preinitiation complex | | Descriptor: | EUKARYOTIC INITIATION FACTOR 3, EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT B, EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT I | | Authors: | des-Georges, A, Dhote, V, Kuhn, L, Hellen, C.U.T, Pestova, T.V, Frank, J, Hashem, Y. | | Deposit date: | 2015-06-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of Mammalian Eif3 in the Context of the 43S Preinitiation Complex.
Nature, 525, 2015
|
|
4YH8
 
 | | Structure of yeast U2AF complex | | Descriptor: | Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ZINC ION | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2015-02-27 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel 3' splice site recognition by the two zinc fingers in the U2AF small subunit.
Genes Dev., 29, 2015
|
|
1N52
 
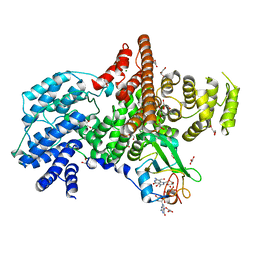 | | Cap Binding Complex | | Descriptor: | 20 kDa nuclear cap binding protein, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, 80 kDa nuclear cap binding protein, ... | | Authors: | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | Deposit date: | 2002-11-04 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
1N54
 
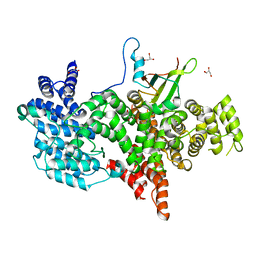 | | Cap Binding Complex m7GpppG free | | Descriptor: | 20 kDa nuclear cap binding protein, 80 kDa nuclear cap binding protein, GLYCEROL | | Authors: | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | Deposit date: | 2002-11-04 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
6PAI
 
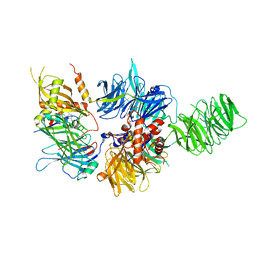 | |
7ONB
 
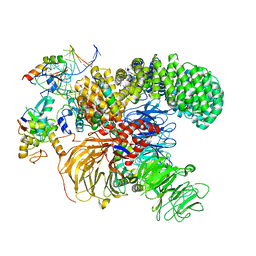 | | Structure of the U2 5' module of the A3'-SSA complex | | Descriptor: | MINX, PHD finger-like domain-containing protein 5A, RNU2, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
7OQB
 
 | | The U2 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | ACT1 pre-mRNA (delta-BS-A), Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing ATP-dependent RNA helicase PRP5, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQE
 
 | | Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A), Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQC
 
 | | The U1 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A), Pre-mRNA-processing factor 39, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
6Q0W
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to Indisulam and RBM39 | | Descriptor: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6Q0R
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to E7820 and RBM39 | | Descriptor: | 3-cyano-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
7PCV
 
 | |
7PY2
 
 | | Structure of pathological TDP-43 filaments from ALS with FTLD | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Arseni, D, Hasegawa, H, Murzin, A.G, Kametani, F, Arai, M, Yoshida, M, Falcon, B. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of pathological TDP-43 filaments from ALS with FTLD.
Nature, 601, 2022
|
|
