2W6N
 
 | |
6CFK
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with D-histidyl-CAM and bound to protein Y (YfiA) at 2.7A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
1EMY
 
 | |
5W1Y
 
 | | SETD8 in complex with a covalent inhibitor | | Descriptor: | 2-(4-methylpiperazin-1-yl)-3-(phenylsulfanyl)naphthalene-1,4-dione, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Yu, W, Li, Y, Blum, G, Luo, M, Pittella-Silva, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-05 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETD8 in complex with a covalent inhibitor
to be published
|
|
9MMR
 
 | | Cryo-EM structure of CRAF/MEK1/14-3-3 complex (open monomer conformation, CRAF Y340D/Y341D mutant) | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Jang, D.M, Jeon, H, Eck, M.J. | | Deposit date: | 2024-12-20 | | Release date: | 2025-09-10 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of CRAF/MEK1/14-3-3 complexes in autoinhibited and open-monomer states reveal features of RAF regulation.
Nat Commun, 16, 2025
|
|
1A29
 
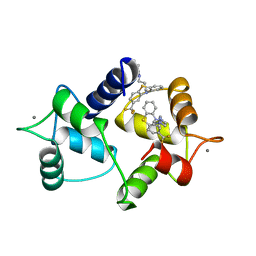 | | CALMODULIN COMPLEXED WITH TRIFLUOPERAZINE (1:2 COMPLEX) | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, CALMODULIN | | Authors: | Bocskei, Zs, Harmat, V, Vertessy, B.G, Ovadi, J, Naray-Szabo, G. | | Deposit date: | 1998-01-19 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Simultaneous binding of drugs with different chemical structures to Ca2+-calmodulin: crystallographic and spectroscopic studies.
Biochemistry, 37, 1998
|
|
3Q8G
 
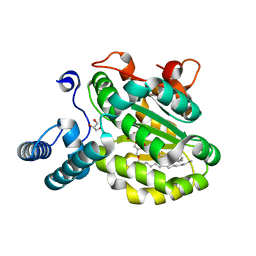 | | Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CRAL-TRIO domain-containing protein YKL091C, GLYCEROL | | Authors: | Ortlund, E.A, Schaaf, G, Bankaitis, V.A. | | Deposit date: | 2011-01-06 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution.
Mol.Biol.Cell, 22, 2011
|
|
4C1A
 
 | |
4YVH
 
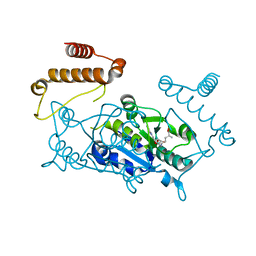 | |
2VO5
 
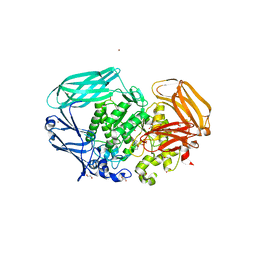 | | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases | | Descriptor: | (1R,4R,5R,7R,8R)-2-Benzyl-5-hydroxymethyl-2-aza-bicyclo[2.2.2]octane-4,7,8-triol, 1,2-ETHANEDIOL, BETA-MANNOSIDASE, ... | | Authors: | Tailford, L.E, Offen, W.A, Smith, N.L, Dumon, C, Moreland, C, Gratien, J, Heck, M.P, Stick, R.V, Bleriot, Y, Vasella, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Evidence for a Boat-Like Transition State in Beta-Mannosidases.
Nat.Chem.Biol., 4, 2008
|
|
6KEO
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in 17beta-estradiol-bound form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ESTRADIOL, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
4YVU
 
 | | Crystal structure of CotA native enzyme in the acid condition, PH5.6 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2015-03-20 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel binding site
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3ZCI
 
 | |
9MDQ
 
 | | Crystal Structure of SARS-CoV-2 Omicron Main Protease (Mpro) Complex with Azapeptide Inhibitor 20a | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-{2-(dichloroacetyl)-2-[(2-oxo-1,2-dihydropyridin-3-yl)methyl]hydrazin-1-yl}-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Vishwakarma, J, Taylor, A.B, Harris, R.S. | | Deposit date: | 2024-12-05 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Azapeptide-Based SARS-CoV-2 Main Protease Inhibitors: Design, Synthesis, Enzyme Inhibition, Structural Determination, and Antiviral Activity.
J.Med.Chem., 68, 2025
|
|
4Y6G
 
 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor in the alpha-site and beta-site. | | Descriptor: | 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2015-02-13 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
3WY3
 
 | | Crystal structure of alpha-glucosidase mutant D202N in complex with glucose and glycerol | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5R51
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N14003a | | Descriptor: | 4-(1,2,3-thiadiazol-4-yl)phenyl ethylcarbamate, DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3K9O
 
 | | The crystal structure of E2-25K and UBB+1 complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of E2-25K/UBB+1 Interaction for Neurotoxicity of Alzheimer Disease by Proteasome Inhibition
To be Published
|
|
5VPA
 
 | | Transcription factor FosB/JunD bZIP domain | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
3GEP
 
 | |
3QGM
 
 | | p-nitrophenyl phosphatase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, p-nitrophenyl phosphatase (Pho2) | | Authors: | Osipiuk, J, Zheng, H, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-01-24 | | Release date: | 2011-02-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p-nitrophenyl phosphatase from Archaeoglobus fulgidus.
To be Published
|
|
1IOM
 
 | |
2XRU
 
 | | AURORA-A T288E COMPLEXED WITH PHA-828300 | | Descriptor: | 3-({[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]CARBONYL}AMINO)-N-[(1R)-1-PHENYLPROPYL]-1H-THIENO[3,2-C]PYRAZOLE-5-CARBOXAMIDE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Bindi, S, Fancelli, D, Alli, C, Berta, D, Bertrand, J.A, Cameron, A.D, Cappella, P, Carpinelli, P, Cervi, G, Croci, W, D'Anello, M, Forte, B, LauraGiorgini, M, Marsiglio, A, Moll, J, Pesenti, E, Pittala, V, Pulici, M, Riccardi-Sirtori, F, Roletto, F, Soncini, C, Storici, P, Varasi, M, Volpi, D, Zugnoni, P, Vianello, P. | | Deposit date: | 2010-09-22 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Thieno[3,2-C]Pyrazoles: A Novel Class of Aurora Inhibitors with Favorable Antitumor Activity.
Bioorg.Med.Chem., 18, 2010
|
|
7FTF
 
 | | Crystal Structure of apo human cyclic GMP-AMP synthase | | Descriptor: | 1,2-ETHANEDIOL, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
5VUA
 
 | | Pim1 Kinase in complex with a benzofuranone inhibitor | | Descriptor: | (2Z)-6-methoxy-7-(piperazin-1-ylmethyl)-2-(1H-pyrrolo[2,3-c]pyridin-3-ylmethylidene)-1-benzofuran-3-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Theoretical Analysis of Activity Cliffs among Benzofuranone-Class Pim1 Inhibitors Using the Fragment Molecular Orbital Method with Molecular Mechanics Poisson-Boltzmann Surface Area (FMO+MM-PBSA) Approach
J Chem Inf Model, 57, 2017
|
|
