3TNH
 
 | | CDK9/cyclin T in complex with CAN508 | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Baumli, S, Hole, A.J, Endicott, J.A. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
1Q9Y
 
 | | CRYSTAL STRUCTURE OF ENTEROBACTERIA PHAGE RB69 GP43 DNA POLYMERASE COMPLEXED WITH 8-OXOGUANOSINE CONTAINING DNA | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-AC(8-OXOGUANOSINE)GGTAAGCAGTCCGCG-3', 5'-GCGGACTGCTTAC(DIDEOXYCYTIDINE)-3', ... | | Authors: | Freisinger, E, Grollman, A.P, Miller, H, Kisker, C. | | Deposit date: | 2003-08-26 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lesion (in)tolerance reveals insights into DNA replication fidelity.
Embo J., 23, 2004
|
|
8PK2
 
 | | Cryo EM structure of the type 1m polymorph of alpha-synuclein | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
5FQO
 
 | |
5FQN
 
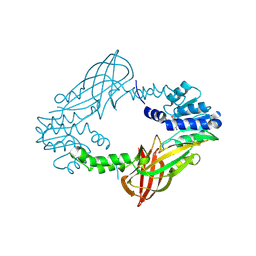 | |
8PIX
 
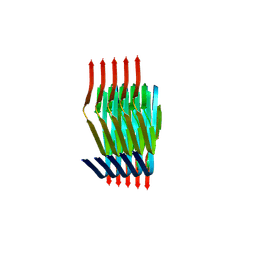 | | Cryo EM structure of the type 3C polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-22 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PJO
 
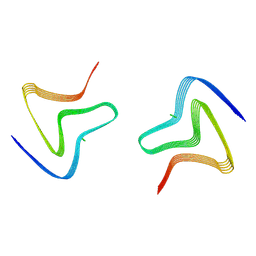 | | Cryo EM structure of the type 3D polymorph of alpha-synuclein E46K mutant at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PK4
 
 | | Cryo EM structure of the type 5A polymorph of alpha-synuclein. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
1HRI
 
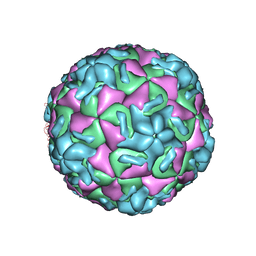 | | STRUCTURE DETERMINATION OF ANTIVIRAL COMPOUND SCH 38057 COMPLEXED WITH HUMAN RHINOVIRUS 14 | | Descriptor: | 1-[6-(2-CHLORO-4-METHYXYPHENOXY)-HEXYL]-IMIDAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Zhang, A, Nanni, R.G, Oren, D.A, Arnold, E. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of antiviral compound SCH 38057 complexed with human rhinovirus 14.
J.Mol.Biol., 230, 1993
|
|
3W0I
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}phenyl)pentan-3-yl]phenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 Receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
3VFZ
 
 | |
3TNW
 
 | | Structure of CDK2/cyclin A in complex with CAN508 | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Baumli, S, Hole, A.J, Endicott, J.A. | | Deposit date: | 2011-09-02 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
3VJS
 
 | | Vitamin D receptor complex with a carborane compound | | Descriptor: | 1-(2-[(S)-2,4-Dihydroxybutoxy]ethyl)-12-(5-ethyl-5-hydroxyheptyl)-1,12-dicarba-closo-dodecaborane, Vitamin D3 receptor, peptide from Mediator of RNA polymerase II transcription subunit 1 | | Authors: | Fujii, S, Masuno, M, Kagechika, H, Nakabayashi, M, Ito, N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Boron Cluster-based Development of Potent Nonsecosteroidal Vitamin D Receptor Ligands: Direct Observation of Hydrophobic Interaction between Protein Surface and Carborane
J.Am.Chem.Soc., 133, 2011
|
|
3VRT
 
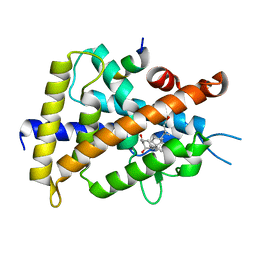 | | VDR ligand binding domain in complex with 2-Mehylidene-19,25,26,27-tetranor-1alpha,24-dihydroxyvitaminD3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-hydroxypentan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yoshimoto, N, Inaba, Y, Itoh, T, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
3VRU
 
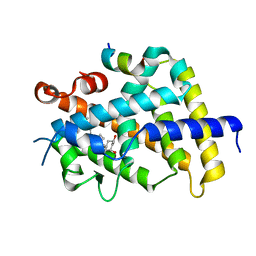 | | VDR ligand binding domain in complex with 2-Methylidene-19,24-dinor-1alpha,25-dihydroxy vitaminD3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-hydroxy-5-methylhexan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Yoshimoto, N, Inaba, Y, Itoh, T, Nakabayashi, M, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
2X5Y
 
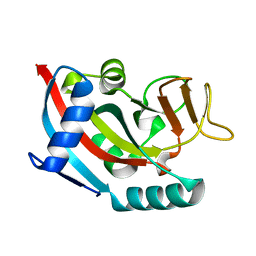 | | Human ZC3HAV1 (ARTD13), C-terminal domain | | Descriptor: | ZINC FINGER CCCH-TYPE ANTIVIRAL PROTEIN 1 | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Kraulis, P, Moche, M, Nordlund, P, Nyman, T, Persson, C, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H. | | Deposit date: | 2010-02-11 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Basis for Lack of Adp-Ribosyltransferase Activity in Poly(Adp-Ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
3VRW
 
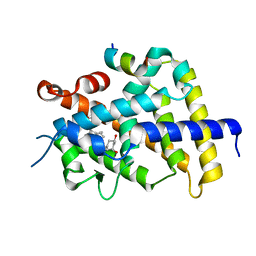 | | VDR ligand binding domain in complex with 22S-Butyl-2-methylidene-26,27-dimethyl-19,24-dinor-1alpha,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3S)-3-butyl-5-ethyl-5-hydroxyheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Yoshimoto, N, Inaba, Y, Itoh, T, Nakabayashi, M, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-16 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
3VRV
 
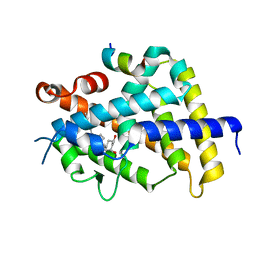 | | VDR ligand binding domain in complex with 2-Methylidene-26,27-dimethyl-19,24-dinor-1alpha,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-ethyl-5-hydroxyheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Yoshimoto, N, Inaba, Y, Itoh, T, Nakabayashi, M, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
3TN8
 
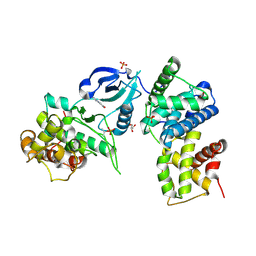 | | CDK9/cyclin T in complex with CAN508 | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Baumli, S, Hole, A.J, Endicott, J.E. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
1Q9X
 
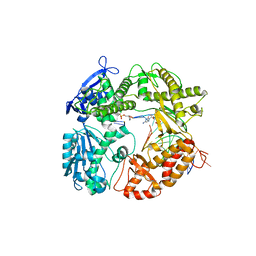 | | Crystal structure of Enterobacteria phage RB69 gp43 DNA polymerase complexed with tetrahydrofuran containing DNA | | Descriptor: | 1',2'-DIDEOXYRIBOFURANOSE-5'-PHOSPHATE, 2',3'-DIDEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Freisinger, E, Grollman, A.P, Miller, H, Kisker, C. | | Deposit date: | 2003-08-26 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Lesion (in)tolerance reveals insights into DNA replication fidelity.
Embo J., 23, 2004
|
|
3WT5
 
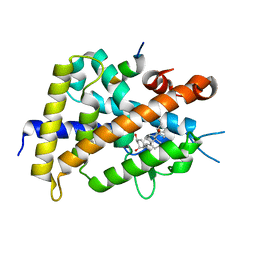 | | A mixed population of antagonist and agonist binding conformers in a single crystal explains partial agonism against vitamin D receptor: Active vitamin D analogues with 22R-alkyl group | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3R)-3-butyl-6-hydroxy-6-methylheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
J.Med.Chem., 57, 2014
|
|
3WTQ
 
 | | Crystal structure of VDR-LBD complexed with 22S-butyl-2-methylidene-19-nor-1a,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3S)-3-butyl-6-hydroxy-6-methylheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Inaba, Y, Itoh, T, Anami, Y, Ikura, T, Ito, N, Yamamoto, K. | | Deposit date: | 2014-04-15 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of VDR-LBD complexed with 22S-butyl-2-methylidene-19-nor-1a,25-dihydroxyvitamin D3
To be Published
|
|
3W0H
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[4-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}phenyl)heptan-4-yl]phenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
3W0J
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[2-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)propan-2-yl]-2-methylphenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 Receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
3W0G
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[2-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}phenyl)propan-2-yl]phenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
