6LD2
 
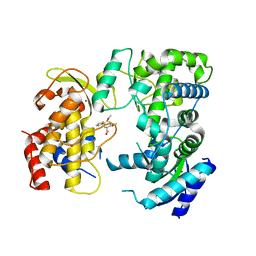 | | Zika NS5 polymerase domain | | Descriptor: | (1S,2S,4S,5R)-2,4-dimethoxy-5-thiophen-2-yl-cyclohexane-1-carboxylic acid, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
5AIC
 
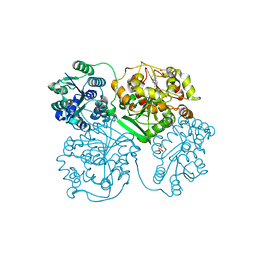 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 7-methyl-2H-1,4-benzothiazin-3(4H)-one, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-02-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5TQE
 
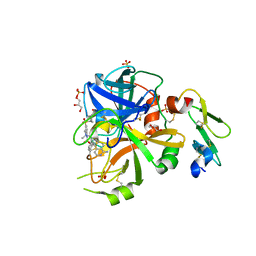 | | Factor VIIa in complex with the inhibitor (5R)-5-[(1-aminoisoquinolin-6-yl)amino]-19-(cyclopropylsulfonyl)-3-methyl-13-oxa-3,15-diazatricyclo[14.3.1.1~6,10~]henicosa-1(20),6(21),7,9,16,18-hexaene-4,14-dione | | Descriptor: | (5R)-5-[(1-aminoisoquinolin-6-yl)amino]-19-(cyclopropylsulfonyl)-3-methyl-13-oxa-3,15-diazatricyclo[14.3.1.1~6,10~]henicosa-1(20),6(21),7,9,16,18-hexaene-4,14-dione, CALCIUM ION, Factor VIIa (Heavy Chain), ... | | Authors: | Wei, A. | | Deposit date: | 2016-10-24 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Novel Meta-Linked Phenylglycine Macrocyclic FVIIa Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
4NRD
 
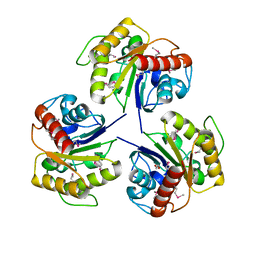 | |
6LG3
 
 | |
5ALO
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 3-BENZYL-3-METHYL-5-(1-METHYLPYRAZOL-4-YL)INDOLIN-2-ONE, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
7FAL
 
 | | Co-crystal Structure of Toxoplasma gondii Prolyl tRNA Synthetase (TgPRS) in complex with T36 and L-pro | | Descriptor: | 4-[(3R)-3-cyano-3-(1-methylcyclopropyl)-2-oxidanylidene-pyrrolidin-1-yl]-N-[[3-fluoranyl-5-(1-methylpyrazol-4-yl)phenyl]methyl]-6-methyl-pyridine-2-carboxamide, PROLINE, Prolyl-tRNA synthetase (ProRS) | | Authors: | Malhotra, N, Mishra, S, Yogavel, M, Sharma, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.219 Å) | | Cite: | Targeting prolyl-tRNA synthetase via a series of ATP-mimetics to accelerate drug discovery against toxoplasmosis.
Plos Pathog., 19, 2023
|
|
5VUA
 
 | | Pim1 Kinase in complex with a benzofuranone inhibitor | | Descriptor: | (2Z)-6-methoxy-7-(piperazin-1-ylmethyl)-2-(1H-pyrrolo[2,3-c]pyridin-3-ylmethylidene)-1-benzofuran-3-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Theoretical Analysis of Activity Cliffs among Benzofuranone-Class Pim1 Inhibitors Using the Fragment Molecular Orbital Method with Molecular Mechanics Poisson-Boltzmann Surface Area (FMO+MM-PBSA) Approach
J Chem Inf Model, 57, 2017
|
|
5AIK
 
 | | Human DYRK1A in complex with LDN-211898 | | Descriptor: | 4-(7-METHOXY-1-(TRIFLUOROMETHYL)-9H-PYRIDO[3,4-B]INDOL-9-yl)butan-1-amine, DYRK1A DUAL-SPECIFICITY TYROSINE-PHOSPHORYLATION REGULATED KINASE 1A, PHOSPHATE ION | | Authors: | Elkins, J.M, Soundararajan, M, Muniz, J.R.C, Cuny, G, Higgins, J, Edwards, A, Bountra, C, Knapp, S. | | Deposit date: | 2015-02-15 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dyrk1A with Ldn-211898
To be published
|
|
3IA6
 
 | |
6F7B
 
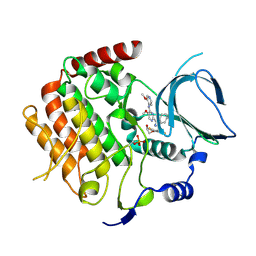 | | Crystal structure of the human Bub1 kinase domain in complex with BAY 1816032 | | Descriptor: | 2-[3,5-bis(fluoranyl)-4-[[3-[5-methoxy-4-[(3-methoxypyridin-4-yl)amino]pyrimidin-2-yl]indazol-1-yl]methyl]phenoxy]ethanol, MAGNESIUM ION, Mitotic checkpoint serine/threonine-protein kinase BUB1 | | Authors: | Holton, S.J, Siemeister, G, Mengel, A, Bone, W, Schroeder, J, Zitzmann-Kolbe, S, Briem, H, Fernandez-Montalvan, A, Prechtl, S, Moenning, U, von Ahsen, O, Johanssen, J, Cleve, A, Puetter, V, Hitchcock, M, von Nussbaum, F, Brands, M, Mumberg, D, Ziegelbauer, K. | | Deposit date: | 2017-12-08 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of BUB1 Kinase by BAY 1816032 Sensitizes Tumor Cells toward Taxanes, ATR, and PARP InhibitorsIn VitroandIn Vivo.
Clin.Cancer Res., 25, 2019
|
|
6KQK
 
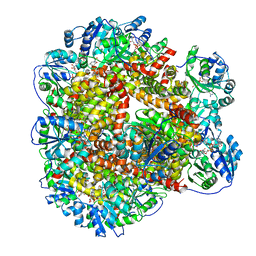 | | 323 K cryoEM structure of Sso-KARI in complex with Mg2+, NADH and CPD | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6B8J
 
 | |
7F9T
 
 | |
5VZ4
 
 | | Receptor-growth factor crystal structure at 2.20 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, GDNF family receptor alpha-like, ... | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2017-05-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15.
Nature, 550, 2017
|
|
3IU2
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90096 | | Descriptor: | (2R)-2-{4-hydroxy-5-methoxy-2-[3-(4-methylpiperazin-1-yl)propyl]phenyl}-3-pyridin-3-yl-1,3-thiazolidin-4-one, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-29 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90096
To be Published
|
|
6KVW
 
 | |
2ANL
 
 | | X-ray crystal structure of the aspartic protease plasmepsin 4 from the malarial parasite plasmodium malariae bound to an allophenylnorstatine based inhibitor | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, plasmepsin IV | | Authors: | Clemente, J.C, Govindasamy, L, Madabushi, A, Fisher, S.Z, Moose, R.E, Yowell, C.A, Hidaka, K, Kimura, T, Hayashi, Y, Kiso, Y, Agbandje-McKenna, M, Dame, J.B, Dunn, B.M, McKenna, R. | | Deposit date: | 2005-08-11 | | Release date: | 2006-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the aspartic protease plasmepsin 4 from the malarial parasite Plasmodium malariae bound to an allophenylnorstatine-based inhibitor.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5AJG
 
 | | Structure of Infrared Fluorescent Protein IFP1.4 AT 1.11 Angstrom resolution | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, BACTERIOPHYTOCHROME | | Authors: | Lafaye, C, Shu, X, Royant, A. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structural Determinants of Improved Fluorescence in a Family of Bacteriophytochrome-Based Infrared Fluorescent Proteins: Insights from Continuum Electrostatic Calculations and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
5ALD
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 5-cyclohexyl-1,3-dihydroindol-2-one, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
4YME
 
 | |
2XOI
 
 | | Functional and Structural Analyses of N-Acylsulfonamide-Linked Dinucleoside Inhibitors of Ribonuclease A | | Descriptor: | (2S,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-N-[[(2S,3S,4R,5R)-5-(2,4-DIOXOPYRIMIDIN-1-YL)-4-HYDROXY-2-(HYDROXYMETHYL)OXOLAN-3-YL]METHYLSULFONYL]-3,4-DIHYDROXY-OXOLANE-2-CARBOXAMIDE, RIBONUCLEASE PANCREATIC | | Authors: | Thiyagarajan, N, Smith, B.D, Raines, R.T, Acharya, K.R. | | Deposit date: | 2010-08-17 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Functional and Structural Analyses of N-Acylsulfonamide-Linked Dinucleoside Inhibitors of Ribonuclease A.
FEBS J., 278, 2011
|
|
5WUK
 
 | | Crystal structure of EED [G255D] in complex with EZH2 peptide and EED226 compound | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase EZH2, N-(furan-2-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Chen, Z. | | Deposit date: | 2016-12-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Split luciferase-based biosensors for characterizing EED binders
Anal. Biochem., 522, 2017
|
|
4YML
 
 | | Crystal structure of Escherichia coli 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3S,4R)-methylthio-DADMe-Immucillin-A | | Descriptor: | (3S,4R)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(methylsulfanyl)methyl]pyrrolidin-3-ol, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, PHOSPHATE ION | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2015-03-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Tight binding enantiomers of pre-clinical drug candidates.
Bioorg.Med.Chem., 23, 2015
|
|
4RHL
 
 | |
