1UYH
 
 | | Human Hsp90-alpha with 9-Butyl-8-(2,5-dimethoxy-benzyl)-2-fluoro-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(2,5-DIMETHOXY-BENZYL)-2-FLUORO-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
3AUA
 
 | | Crystal structure of the quaternary complex-2 of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[ethanoyl(hydroxy)amino]propylphosphonic acid, CALCIUM ION, ... | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
5G57
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-001 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
1UYE
 
 | | Human Hsp90-alpha with 8-(2-chloro-3,4,5-trimethoxy-benzyl)-9-pent-4-ylnyl-9H-purin-6-ylamine | | Descriptor: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
3ZDV
 
 | | Crystal structure of the LecB lectin from Pseudomonas aeruginosa in complex with Methyl 6-(2,4,6-trimethylphenylsulfonylamido)-6-deoxy-alpha-D-mannopyranoside | | Descriptor: | 1,2-ETHANEDIOL, 2,4,6-trimethylbenzenesulfonamide, CALCIUM ION, ... | | Authors: | Hauck, D, Joachim, I, Frommeyer, B, Varrot, A, Philipp, B, MOller, H.M, Imberty, A, Exner, T.E, Titz, A. | | Deposit date: | 2012-11-30 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Discovery of Two Classes of Potent Glycomimetic Inhibitors of Pseudomonas Aeruginosa Lecb with Distinct Binding Modes.
Acs Chem.Biol., 8, 2013
|
|
3CDA
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-{3-(4-fluorophenyl)-1-(1-methylethyl)-4-phenyl-5-[(4-sulfamoylphenyl)carbamoyl]-1H-pyrrol-2-yl}-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
1UY9
 
 | | Human Hsp90-alpha with 8-Benzo[1,3]dioxol-,5-ylmethyl-9-butyl-9H-purin-6-ylamine | | Descriptor: | 8-BENZO[1,3]DIOXOL-,5-YLMETHYL-9-BUTYL-9H-, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1UZ2
 
 | | The Cys121Ser Mutant of Beta-Lactoglobulin | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | McNae, I, Jayat, D, Haertle, T, Holt, C, Sawyer, L. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A recombinant C121S mutant of bovine beta-lactoglobulin is more susceptible to peptic digestion and to denaturation by reducing agents and heating.
Biochemistry, 43, 2004
|
|
3CEJ
 
 | | Human glycogen phosphorylase (tense state) in complex with the allosteric inhibitor AVE2865 | | Descriptor: | 1-{2-[3-(2-Chloro-4,5-difluoro-benzoyl)-ureido]-4-fluoro-phenyl}-piperidine-4-carboxylic acid, Glycogen phosphorylase, liver form, ... | | Authors: | Wendt, K.U, Dreyer, M.K, Anderka, O, Klabunde, T, Loenze, P, Defossa, E, Schmoll, D. | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Thermodynamic characterization of allosteric glycogen phosphorylase inhibitors.
Biochemistry, 47, 2008
|
|
1UYG
 
 | | Human Hsp90-alpha with 8-(2,5-dimethoxy-benzyl)-2-fluoro-9H-purin-6-ylamine | | Descriptor: | 8-(2,5-DIMETHOXY-BENZYL)-2-FLUORO-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
3CHQ
 
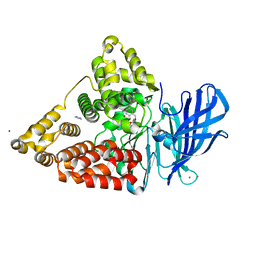 | | Crystal structure of leukotriene a4 hydrolase in complex with N5-[4-(phenylmethoxy)phenyl]-L-glutamine | | Descriptor: | (2S)-2-amino-5-oxo-5-[(4-phenylmethoxyphenyl)amino]pentanoic acid, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
3CG5
 
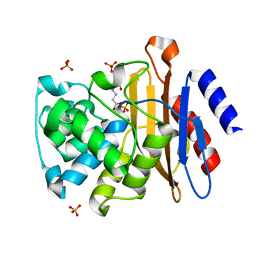 | |
3CHR
 
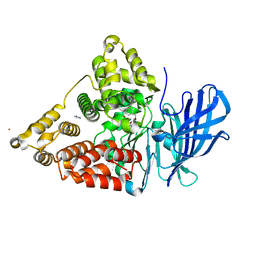 | | Crystal structure of leukotriene A4 hydrolase in complex with 4-amino-N-[4-(phenylmethoxy)phenyl]-butanamide | | Descriptor: | 4-amino-N-[4-(benzyloxy)phenyl]butanamide, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
1US0
 
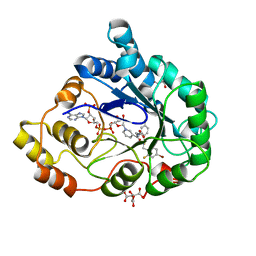 | | Human Aldose Reductase in complex with NADP+ and the inhibitor IDD594 at 0.66 Angstrom | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, IDD594, ... | | Authors: | Howard, E.I, Sanishvili, R, Cachau, R.E, Mitschler, A, Chevrier, B, Barth, P, Lamour, V, Van Zandt, M, Sibley, E, Bon, C, Moras, D, Schneider, T.R, Joachimiak, A, Podjarny, A. | | Deposit date: | 2003-11-16 | | Release date: | 2004-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.66 Å) | | Cite: | Ultrahigh Resolution Drug Design I: Details of Interactions in Human Aldose Reductase-Inhibitor Complex at 0.66 A.
Proteins, 55, 2004
|
|
5HYS
 
 | | Structure of IgE complexed with omalizumab | | Descriptor: | Epididymis luminal protein 214, Ig epsilon chain C region, SULFATE ION, ... | | Authors: | Pennington, L.F, Tarchevskaya, S.S, Sathiyamoorthy, K, Jardetzky, T.S. | | Deposit date: | 2016-02-01 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of omalizumab therapy and omalizumab-mediated IgE exchange.
Nat Commun, 7, 2016
|
|
3AFG
 
 | | Crystal structure of ProN-Tk-SP from Thermococcus kodakaraensis | | Descriptor: | CALCIUM ION, Subtilisin-like serine protease | | Authors: | Foophow, T, Tanaka, S, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a subtilisin homologue, Tk-SP, from Thermococcus kodakaraensis: requirement of a C-terminal beta-jelly roll domain for hyperstability.
J.Mol.Biol., 400, 2010
|
|
7EOW
 
 | |
3ALQ
 
 | | Crystal structure of TNF-TNFR2 complex | | Descriptor: | COBALT (II) ION, Tumor necrosis factor, Tumor necrosis factor receptor superfamily member 1B | | Authors: | Mukai, Y, Nakamura, T, Yamagata, Y, Tsutsumi, Y. | | Deposit date: | 2010-08-06 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Solution of the Structure of the TNF-TNFR2 Complex
Sci.Signal., 3, 2010
|
|
7FAY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with (R)-1a | | Descriptor: | (2~{R})-~{N}-[(1~{R})-2-(~{tert}-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-~{N}-(4-~{tert}-butylphenyl)-2-oxidanyl-propanamide, 3C-like proteinase | | Authors: | Zeng, R, Quan, B.X, Liu, X.L, Lei, J. | | Deposit date: | 2021-07-08 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An orally available M pro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron.
Nat Microbiol, 7, 2022
|
|
3ANV
 
 | | Crystal structure of D-serine dehydratase from chicken kidney (2,3-DAP complex) | | Descriptor: | 3-amino-D-alanine, CHLORIDE ION, D-serine dehydratase, ... | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney
J.Biol.Chem., 286, 2011
|
|
7FAZ
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Y180 | | Descriptor: | (2~{R})-~{N}-dibenzofuran-3-yl-~{N}-[(1~{R})-2-[[(1~{S})-1-(4-fluorophenyl)ethyl]amino]-2-oxidanylidene-1-pyridin-3-yl-ethyl]-2-oxidanyl-propanamide, 3C-like proteinase, SODIUM ION | | Authors: | Zeng, R, Quan, B.X, Liu, X.L, Lei, J. | | Deposit date: | 2021-07-08 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An orally available M pro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron.
Nat Microbiol, 7, 2022
|
|
2XE1
 
 | |
3ANU
 
 | | Crystal structure of D-serine dehydratase from chicken kidney | | Descriptor: | CHLORIDE ION, D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-06-15 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney
J.Biol.Chem., 286, 2011
|
|
2EUF
 
 | |
1TU7
 
 | |
