8SVE
 
 | |
8SSI
 
 | | Structure of Burkholderia pseudomallei deubiquitinase TssM in complex with ubiquitin | | Descriptor: | Deubiquitinase TssM, GLYCEROL, Ubiquitin, ... | | Authors: | Szczesna, M, Pruneda, J.N, Thurston, T.L.M. | | Deposit date: | 2023-05-08 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial esterases reverse lipopolysaccharide ubiquitylation to block host immunity.
Cell Host Microbe, 32, 2024
|
|
8THU
 
 | |
8T9F
 
 | |
8T9H
 
 | |
8GVL
 
 | | PTPN21 FERM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8GWH
 
 | |
8GVV
 
 | | PTPN21 PTP domain C1108S mutant | | Descriptor: | IODIDE ION, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-15 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8GXE
 
 | | PTPN21 FERM PTP complex | | Descriptor: | CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
8TLD
 
 | | Structure of the IL-5 Signaling Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit beta, Interleukin-5, ... | | Authors: | Caveney, N.A, Garcia, K.C. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-10 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the interleukin-5 receptor complex exemplifies the organizing principle of common beta cytokine signaling.
Mol.Cell, 84, 2024
|
|
8GO8
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 | | Descriptor: | Beta-arrestin-1, C5a anaphylatoxin chemotactic receptor 1, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8GOC
 
 | | Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human Vasopressin V2 receptor, V2R | | Descriptor: | Beta-arrestin-2, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8GP3
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8GOO
 
 | | Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 | | Descriptor: | Beta-arrestin-2, C5a anaphylatoxin chemotactic receptor 1, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8TN7
 
 | | The Crystal Structure of a human monoclonal antibody (aAb), termed TG10, complexed with a disaccharide | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SULFATE ION, ... | | Authors: | Li, M, Wlodawer, A, Temme, S, Gildersleeve, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Insights into biofilm architecture and maturation enable improved clinical strategies for exopolysaccharide-targeting therapeutics.
Cell Chem Biol, 31, 2024
|
|
8TN5
 
 | | The Crystal Structure of a human monoclonal antibody (aAb), termed TG10, complexed with a GlcNH2 | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SULFATE ION, ... | | Authors: | Li, M, Wlodawer, A, Temme, S, Gildersleeve, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Insights into biofilm architecture and maturation enable improved clinical strategies for exopolysaccharide-targeting therapeutics.
Cell Chem Biol, 31, 2024
|
|
8TN4
 
 | | The Crystal Structure of a human monoclonal antibody (aAb), termed TG10, used to study poly-N-acetyl-glucosamine broadly expressed in biofilm-forming pathogenclonal antibody | | Descriptor: | SODIUM ION, SULFATE ION, TG10, ... | | Authors: | Li, M, Wlodawer, A, Temme, S, Gildersleeve, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into biofilm architecture and maturation enable improved clinical strategies for exopolysaccharide-targeting therapeutics.
Cell Chem Biol, 31, 2024
|
|
8GAU
 
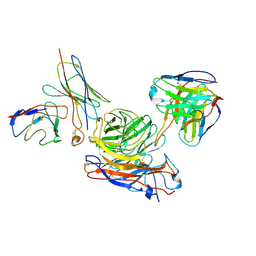 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8GAT
 
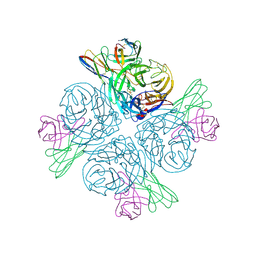 | | Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), based on consensus cryo-EM map with only Fab 1G01 resolved | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 1G01, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8GAV
 
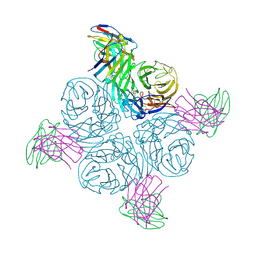 | | Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab NDS.3, heavy chain, ... | | Authors: | Tsybovsky, Y, Lederhofer, J, Kwong, P.D, Kanekiyo, M. | | Deposit date: | 2023-02-23 | | Release date: | 2024-02-28 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Protective human monoclonal antibodies target conserved sites of vulnerability on the underside of influenza virus neuraminidase.
Immunity, 57, 2024
|
|
8I0Z
 
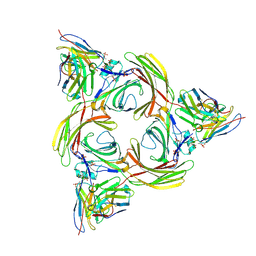 | | Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 (Local refine) | | Descriptor: | Beta-arrestin-2, C5a anaphylatoxin chemotactic receptor 1, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8I0Q
 
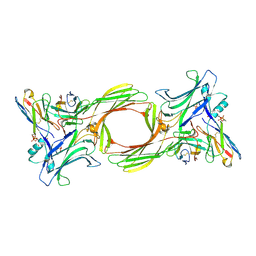 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 (Local refine) | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-11 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8I10
 
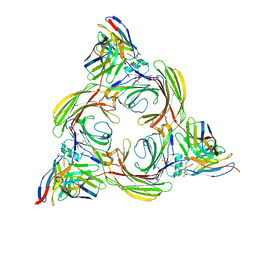 | | Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human Vasopressin V2 receptor, V2R (Local refine) | | Descriptor: | Beta-arrestin-2, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
7UP8
 
 | |
8UK1
 
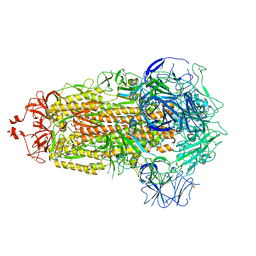 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD-down Spike Protein Trimer consensus (S-RRAR-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-10-11 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
