9D52
 
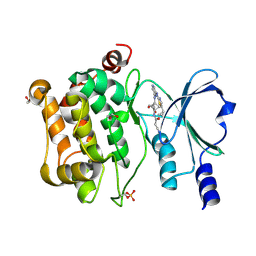 | | Structure of PAK4 in complex with compound 18 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyano-N-[3-({6-[(5-cyclopropyl-1,3-thiazol-2-yl)amino]pyrazin-2-yl}amino)bicyclo[1.1.1]pentan-1-yl]acetamide, Serine/threonine-protein kinase PAK 4 | | Authors: | Boone, C, Suto, R, Olland, A. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 127, 2025
|
|
3II5
 
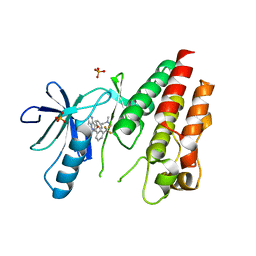 | | The Complex of wild-type B-RAF with Pyrazolo pyrimidine inhibitor | | Descriptor: | B-Raf proto-oncogene serine/threonine-protein kinase, N-[3-(3-{4-[(dimethylamino)methyl]phenyl}pyrazolo[1,5-a]pyrimidin-7-yl)phenyl]-3-(trifluoromethyl)benzamide, PHOSPHATE ION | | Authors: | Xu, W, Breger, D, Torres, N, Dutia, M, Powell, D, Ciszewski, G. | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Non-hinge-binding pyrazolo[1,5-a]pyrimidines as potent B-Raf kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6ECO
 
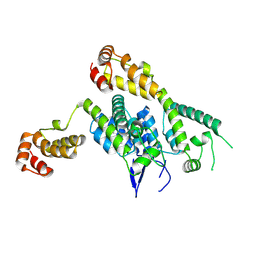 | | Hexamer-2-Foldon HIV-1 capsid platform | | Descriptor: | HIV-1 capsid platform protein | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-08 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
8SAV
 
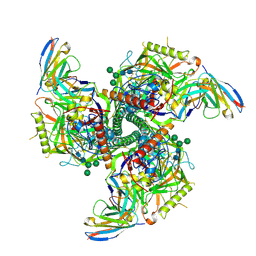 | | CryoEM structure of VRC01-CH848.0526.25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.0526.25 gp120, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAQ
 
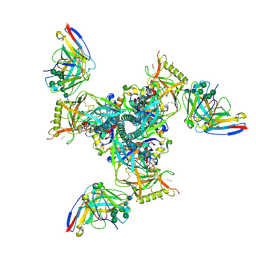 | | CryoEM structure of DH270.6-CH848.0526.25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.0526.25 gp120, CH848.0526.25 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
3EM4
 
 | | Crystal structure of atazanavir (ATV) in complex with I50L/A71V drug-resistant HIV-1 protease | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabalan, M, King, N, Royer, C, Schiffer, C. | | Deposit date: | 2008-09-23 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Structural studies on atazanavir-specific I50L drug-resistant HIV-1 protease mutant
To be Published
|
|
6QJE
 
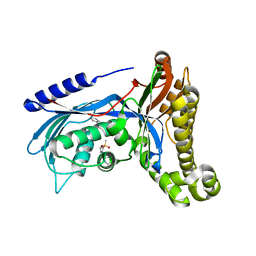 | | Structure of human galactokinase 1 bound with 4-{[2-(Methylsulfonyl)-1H-imidazol-1-yl]methyl}-1,3-thiazole | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, 4-[(2-methylsulfonylimidazol-1-yl)methyl]-1,3-thiazole, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 4-{[2-(Methylsulfonyl)-1H-imidazol-1-yl]methyl}-1,3-thiazole
To Be Published
|
|
5CU8
 
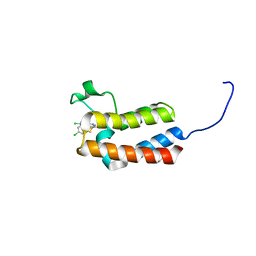 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 2-Amino-6-chlorobenzothiazole (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-1,3-benzothiazol-2-amine, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 2-Amino-6-chlorobenzothiazole (SGC - Diamond I04-1 fragment screening)
To be published
|
|
5CUE
 
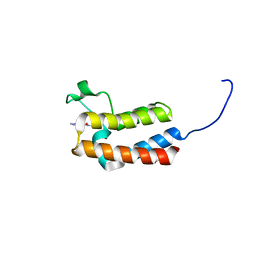 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with AGN-PC-04G0SS (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N,1-dimethyl-1H-pyrazole-4-carboxamide, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with AGN-PC-04G0SS (SGC - Diamond I04-1 fragment screening)
To be published
|
|
3TU9
 
 | |
4LGO
 
 | |
4LBA
 
 | |
6CNX
 
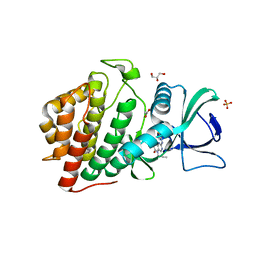 | | Crystal Structure of the Human vaccinia-related kinase 1 (VRK1) bound to an N-propynyl-N-isopentyl-dihydropteridin inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-8-(3-methylbutyl)-5-(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, (7S)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-8-(3-methylbutyl)-5-(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, ACETATE ION, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-09 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Vaccinia-Related Kinase 1 (VRK1) bound to a N-propynyl-N-isopentil-dihydropteridine inhibitor
To Be Published
|
|
4Z35
 
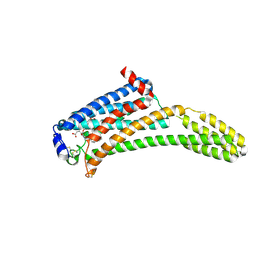 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-9910539 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 3-{1-[(2S,3S)-3-(4-acetyl-3,5-dimethoxyphenyl)-2-(2,3-dihydro-1H-inden-2-ylmethyl)-3-hydroxypropyl]-4-(methoxycarbonyl)-1H-pyrrol-3-yl}propanoic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
3UCG
 
 | |
6ALA
 
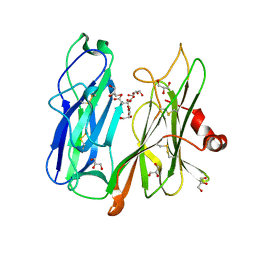 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | Descriptor: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6D3Y
 
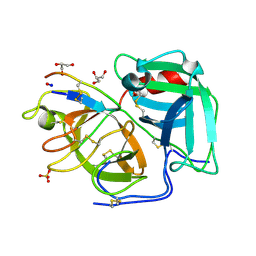 | |
5CQ3
 
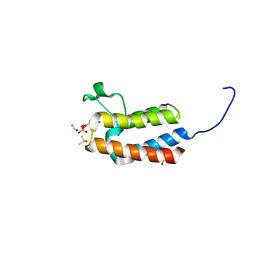 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 6-Hydroxypicolinic acid (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 6-hydroxypyridine-2-carboxylic acid, Bromodomain adjacent to zinc finger domain protein 2B, ... | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 6-Hydroxypicolinic acid (SGC - Diamond I04-1 fragment screening)
To be published
|
|
5CUD
 
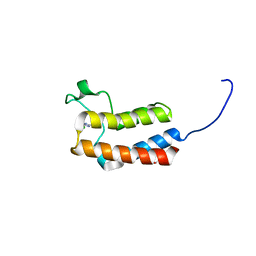 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 6-CHLOROPURINE (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-9H-purine, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 6-CHLOROPURINE (SGC - Diamond I04-1 fragment screening)
To be published
|
|
3IIT
 
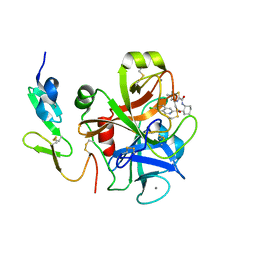 | | Factor XA in complex with a cis-1,2-diaminocyclohexane derivative | | Descriptor: | 7-chloro-N-[(1S,2R,4S)-4-(dimethylcarbamoyl)-2-{[(5-methyl-5,6-dihydro-4H-pyrrolo[3,4-d][1,3]thiazol-2-yl)carbonyl]amino}cyclohexyl]isoquinoline-3-carboxamide, Activated factor Xa heavy chain, CALCIUM ION, ... | | Authors: | Suzuki, M. | | Deposit date: | 2009-08-03 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, synthesis, and SAR of cis-1,2-diaminocyclohexane derivatives as potent factor Xa inhibitors. Part II: exploration of 6-6 fused rings as alternative S1 moieties.
Bioorg.Med.Chem., 17, 2009
|
|
8I4D
 
 | | X-ray structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
7BYV
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A E208Q with beta-1,3-galactotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Galactan 1,3-beta-galactosidase, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
3EL1
 
 | | Crystal Structure of wild-type HIV protease in complex with the inhibitor, Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
4PUX
 
 | |
4Z36
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-3080573 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 1-(4-{[(2S,3R)-2-(2,3-dihydro-1H-inden-2-yloxy)-3-(3,5-dimethoxy-4-methylphenyl)-3-hydroxypropyl]oxy}phenyl)cyclopropan ecarboxylic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
