5MAZ
 
 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PA14 IN COMPLEX WITH 3-Thiophenesulfonamide-2,5-dimethyl-N-methyl-beta-L-fucopyranoside | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Sommer, R, Imberty, A, Titz, A, Varrot, A. | | Deposit date: | 2016-11-07 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of a new class of C-glycoside based inhibitors of the virulence factor LecB from Pseudomonas aeruginosa
To Be Published
|
|
6MVK
 
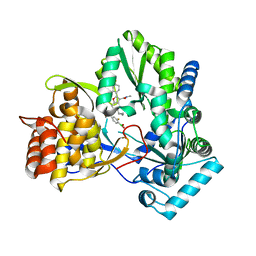 | | HCV NS5B 1b N316 bound to Compound 18 | | Descriptor: | (4-{(4S)-3-[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl]-2-oxo-1,3-oxazolidin-4-yl}-2-fluorophenyl)boronic acid, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
8FKR
 
 | |
6QB9
 
 | | Structure of an anti-Mcl1 scFv | | Descriptor: | L(+)-TARTARIC ACID, scFv55 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4YIS
 
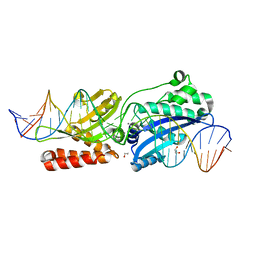 | | Crystal Structure of LAGLIDADG Meganuclease I-CpaMI Bound to Uncleaved DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (28-MER), ... | | Authors: | Hallinan, J.P, Kaiser, B.K, Stoddard, B.L. | | Deposit date: | 2015-03-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity.
Structure, 24, 2016
|
|
8C1E
 
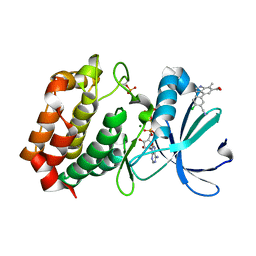 | | Aurora A kinase in complex with TPX2-inhibitor 9 | | Descriptor: | 4-(4-chloranyl-3-cyano-phenyl)-7-methyl-1~{H}-indole-6-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Fischer, G, Rocaboy, M, Blaszczyk, B, Moschetti, T, Wang, X, Scott, D.E, Coyne, A.G, Dagostin, C, Rooney, T, Bayly, A, Feng, J, Asteian, A, Alcaide-Lopez, A, Stockwell, S, Skidmore, J, Venkitaraman, A.R, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Selective Aurora A-TPX2 Interaction Inhibitors Have In Vivo Efficacy as Targeted Antimitotic Agents.
J.Med.Chem., 67, 2024
|
|
7QI8
 
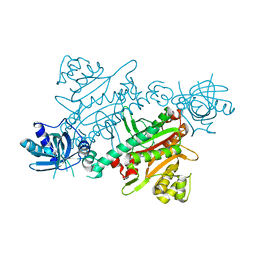 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE AND INHIBITOR | | Descriptor: | 2-azanyl-6-[(1~{S},7~{S})-2,2-bis(fluoranyl)-7-oxidanyl-cycloheptyl]-4-methoxy-7~{H}-pyrrolo[3,4-d]pyrimidin-5-one, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
6VUW
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT368 | | Descriptor: | (7R)-7-methyl-2-({[(3R)-1-methylpiperidin-3-yl]methyl}sulfanyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-amine, GLYCEROL, N-acetyltransferase Eis, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
8OGQ
 
 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry B06 | | Descriptor: | (1S)-1-(4-nitrophenyl)ethan-1-ol, Cyclic di-AMP synthase CdaA, MAGNESIUM ION | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-27 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
7QH8
 
 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE | | Descriptor: | GLYCEROL, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-10 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
7P39
 
 | | 4,6-alpha-glucanotransferase GtfB from Limosilactobacillus reuteri NCC 2613 complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, Dextransucrase, ... | | Authors: | Pijning, T, te Poele, E, Gangoiti, J, Boerner, T, Dijkhuizen, L. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into Broad-Specificity Starch Modification from the Crystal Structure of Limosilactobacillus Reuteri NCC 2613 4,6-alpha-Glucanotransferase GtfB.
J.Agric.Food Chem., 69, 2021
|
|
8OZ8
 
 | | Crystal Structure of an Hydroxynitrile lyase variant (H96A) from Granulicella tundricola | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, BROMIDE ION, ... | | Authors: | Bento, I, Coloma, J, Hagedoorn, P.-L, Hanefeld, U. | | Deposit date: | 2023-05-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Can a Hydroxynitrile Lyase Catalyze an Oxidative Cleavage?
Acs Catalysis, 13, 2023
|
|
7ANB
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-acetylgalactosamine-6-sulfatase, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
6N5G
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 2 | | Descriptor: | 4-[(quinolin-3-yl)methyl]-N-[4-(trifluoromethoxy)phenyl]piperidine-1-carboxamide, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
5JWH
 
 | | Apo structure | | Descriptor: | 1,2-ETHANEDIOL, NS3 helicase | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-12 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
8U1P
 
 | | Local refinement of Plasmodium falciparum gametocyte surface protein Pfs48/45 Domains 1 and 2 in complex with neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gametocyte surface protein P45/48, ... | | Authors: | Kucharska, I, Hailemariam, S, Ivanochko, D, Rubinstein, J, Julien, J.P. | | Deposit date: | 2023-09-01 | | Release date: | 2025-03-12 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural elucidation of full-length Pfs48/45 in complex with potent monoclonal antibodies isolated from a naturally exposed individual.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6QEE
 
 | |
8RB1
 
 | | The crystal structure of DNA-bound human MutSbeta (MSH2_K675R/MSH3_K902R) in the canonical mismatch bound conformation with ADP bound in MSH2 and MSH3 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thomsen, M, Neudegger, T, Thieulin-Pardo, G, Blaesse, M, Costanzi, E, Steinbacher, S, Plotnikov, N.V, Dominguez, C, Iyer, R.R, Wilkinson, H.A, Monteagudo, E, Haque, T.S, Prasad, B.C, Finley, M, Boudet, J, Vogt, T.F, Felsenfeld, D.P. | | Deposit date: | 2023-12-01 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | The crystal structure of DNA-bound human MutSbeta (MSH2/MSH3) in the canonical mismatch bound conformation with ADP bound in MSH2 and MSH3
To Be Published
|
|
6YQ0
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3-oxidanyl-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
8OHL
 
 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry H09 | | Descriptor: | (2~{R})-2-fluoranyl-2-(4-fluoranyl-1,2,4$l^{4}-triazacyclopenta-2,4-dien-1-yl)-1-phenyl-ethanone, Cyclic di-AMP synthase CdaA, MAGNESIUM ION | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-04-03 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
7PD3
 
 | | Structure of the human mitoribosomal large subunit in complex with NSUN4.MTERF4.GTPBP7 and MALSU1.L0R8F8.mt-ACP | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Chandrasekaran, V, Desai, N, Burton, N.O, Yang, H, Price, J, Miska, E.A, Ramakrishnan, V. | | Deposit date: | 2021-08-04 | | Release date: | 2021-11-17 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualizing formation of the active site in the mitochondrial ribosome.
Elife, 10, 2021
|
|
8UL1
 
 | |
9KNZ
 
 | | ERDRP-0519-bound Nipah virus L-P complex | | Descriptor: | 2-methyl-~{N}-[4-[(2~{S})-2-(2-morpholin-4-ylethyl)piperidin-1-yl]sulfonylphenyl]-5-(trifluoromethyl)pyrazole-3-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Wang, Y.R, Zhang, H.Q. | | Deposit date: | 2024-11-19 | | Release date: | 2025-07-16 | | Last modified: | 2025-09-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the measles virus polymerase complex with non-nucleoside inhibitors and mechanism of inhibition.
Cell, 188, 2025
|
|
6W90
 
 | | De novo designed NTF2 fold protein NT-9 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, NTF2 fold protein loop-helix-loop design NT-9 | | Authors: | Thompson, M.C, Pan, X, Liu, L, Fraser, J.S, Kortemme, T. | | Deposit date: | 2020-03-21 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Expanding the space of protein geometries by computational design of de novo fold families.
Science, 369, 2020
|
|
6H91
 
 | |
