7RZ4
 
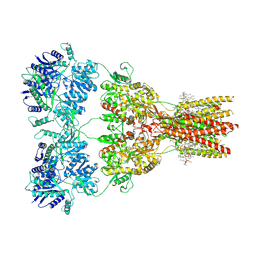 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK 200775 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, ... | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
5XL3
 
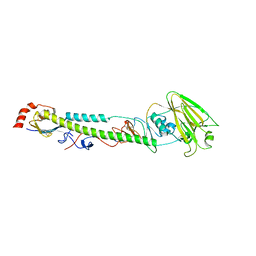 | | Complex structure of H4 hemagglutinin from avian influenza H4N6 virus with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation by Influenza A Virus Hemagglutinin H4
Cell Rep, 20, 2017
|
|
4V7J
 
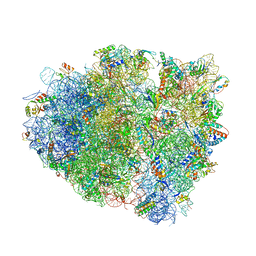 | | Structure of RelE nuclease bound to the 70S ribosome (precleavage state) | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | Deposit date: | 2009-11-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
5DH9
 
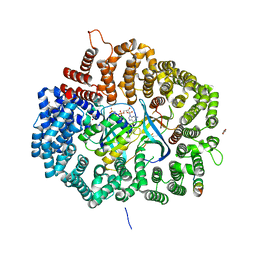 | |
5DHT
 
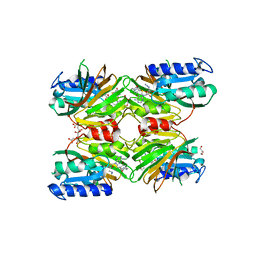 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a novel inhibitor | | Descriptor: | 5'-azido-8-[(2-{[2-(3-bromophenyl)ethyl]amino}-2-oxoethyl)sulfanyl]-5'-deoxyadenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Paoletti, J, Assairi, L, Huteau, V, Pochet, S, Labesse, G. | | Deposit date: | 2015-08-31 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | 8-Thioalkyl-adenosine derivatives inhibit Listeria monocytogenes NAD kinase through a novel binding mode.
Eur.J.Med.Chem., 124, 2016
|
|
7FCP
 
 | |
4V8O
 
 | | Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3 | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jin, H, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2011-07-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal Structure of the Hybrid State of Ribosome in Complex with the Guanosine Triphosphatase Release Factor 3.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7RPI
 
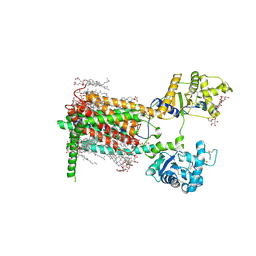 | | Cryo-EM structure of murine Dispatched 'T' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
5DID
 
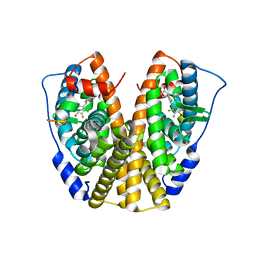 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a difluoro-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-5-(2,3-difluoro-4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-5-(2,3-difluoro-4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4V7K
 
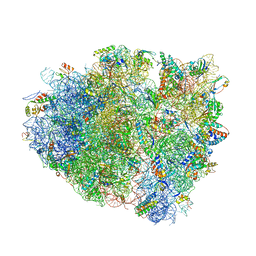 | | Structure of RelE nuclease bound to the 70S ribosome (postcleavage state) | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | Deposit date: | 2009-11-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5D8P
 
 | | 2.35A resolution structure of iron bound BfrB (wild-type, C2221 form) from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
5D9D
 
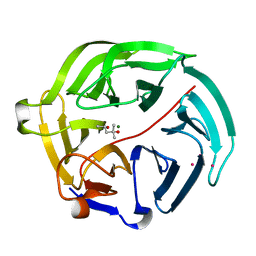 | | Luciferin-regenerating enzyme solved by SAD using synchrotron radiation at room temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
5XWS
 
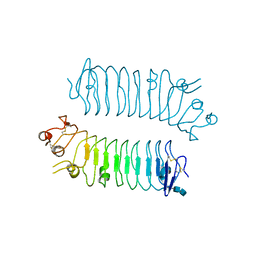 | | Crystal structure of SALM5 LRR-Ig | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat and fibronectin type-III domain-containing protein 5 | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.084 Å) | | Cite: | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
7S11
 
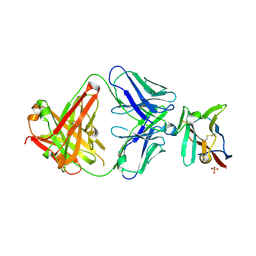 | | Crystal structure of Fab in complex with mouse CD96 monomer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-08-31 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Antibody blockade of CD96 by distinct molecular mechanisms.
Mabs, 13, 2021
|
|
4V6Z
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in classic pre-translocation state (pre1b) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V7D
 
 | | Structure of the Ribosome with Elongation Factor G Trapped in the Pre-Translocation State (pre-translocation 70S*tRNA*EF-G structure) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Brilot, A.F, Korostelev, A.A, Ermolenko, D.N, Grigorieff, N. | | Deposit date: | 2013-11-21 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the ribosome with elongation factor G trapped in the pretranslocation state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5DIT
 
 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand (1R)-3-(3,4-dimethoxyphenyl)-1-f3-[2-(morpholin-4-yl)ethoxy]phenylgpropyl(2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate | | Descriptor: | (1R)-3-(3,4-dimethoxyphenyl)-1-{3-[2-(morpholin-4-yl)ethoxy]phenyl}propyl (2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Affinity Relationship Analysis of Selective FKBP51 Ligands.
J.Med.Chem., 58, 2015
|
|
4V8Q
 
 | | Complex of SmpB, a tmRNA fragment and EF-Tu-GDP-Kirromycin with the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Neubauer, C, Gillet, R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2011-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Decoding in the absence of a codon by tmRNA and SmpB in the ribosome.
Science, 335, 2012
|
|
7F44
 
 | | Crystal structure of Moraxella catarrhalis enoyl-ACP-reductase (FabI) in complex with the cofactor NAD | | Descriptor: | CALCIUM ION, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Katiki, M, Neetu, N, Pratap, S, Kumar, P. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Biochemical and structural basis for Moraxella catarrhalis enoyl-acyl carrier protein reductase (FabI) inhibition by triclosan and estradiol.
Biochimie, 198, 2022
|
|
5DJ5
 
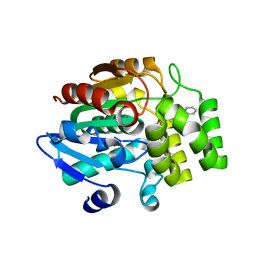 | | Crystal structure of rice DWARF14 in complex with synthetic strigolactone GR24 | | Descriptor: | (3E,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methylidene)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, Probable strigolactone esterase D14 | | Authors: | Zhou, X.E, Zhao, L.-H, Yi, W, Wu, Z.-S, Liu, Y, Kang, Y, Hou, L, de Waal, P.W, Li, S, Jiang, Y, Melcher, K, Xu, H.E. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.
Cell Res., 25, 2015
|
|
5DAN
 
 | |
4V8A
 
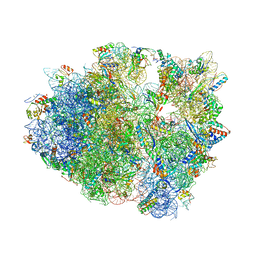 | | The structure of thermorubin in complex with the 70S ribosome from Thermus thermophilus. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D, Johnson, F.A, Steitz, T.A. | | Deposit date: | 2011-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The antibiotic thermorubin inhibits protein synthesis by binding to inter-subunit bridge b2a of the ribosome.
J.Mol.Biol., 416, 2012
|
|
5DAT
 
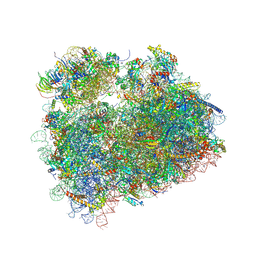 | | Complex of yeast 80S ribosome with hypusine-containing eIF5A | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
4V8R
 
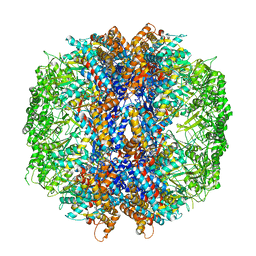 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Kalisman, N, Schroder, G.F, Levitt, M. | | Deposit date: | 2012-03-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
