8R5C
 
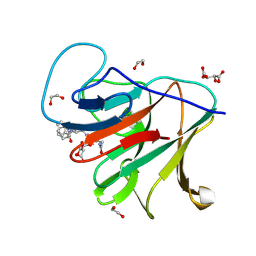 | | Crystal structure of human TRIM7 PRYSPRY domain bound to (2-(1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine | | Descriptor: | (2~{S})-5-azanyl-5-oxidanylidene-2-[[(2~{S})-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)-3-phenyl-propanoyl]amino]pentanoic acid, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, ... | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
6ADS
 
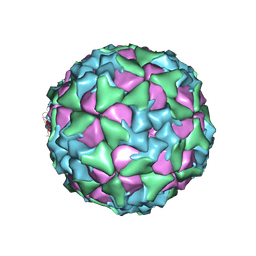 | | Structure of Seneca Valley Virus in acidic conditions | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8EZ1
 
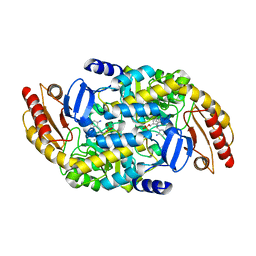 | | Human Ornithine Aminotransferase (hOAT) co-crystallized with its inactivator 3-Amino-4-fluorocyclopentenecarboxylic Acid | | Descriptor: | (1R,3S,4Z)-3-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-4-iminocyclopentane-1-carboxylic acid, (3E,4E)-4-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-3-iminocyclopent-1-ene-1-carboxylic acid, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Shen, S, Silverman, R, Liu, D. | | Deposit date: | 2022-10-30 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural and Mechanistic Basis for the Inactivation of Human Ornithine Aminotransferase by (3 S ,4 S )-3-Amino-4-fluorocyclopentenecarboxylic Acid.
Molecules, 28, 2023
|
|
8XWU
 
 | | Crystal structure of human 8-oxoguanine glycosylase K249H mutant bound to the reaction intermediate derived from the crystal soaked into the solution at pH 4.0 under 277 K for 24 hourss | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*CP*A)-3'), DNA (5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*TP*AP*CP*C)-3'), ... | | Authors: | Unno, M, Koga, M, Minowa, N, Komuro, S, Tanaka, Y. | | Deposit date: | 2024-01-16 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Capturing a glycosylase reaction intermediate in DNA repair by freeze-trapping of a pH-responsive hOGG1 mutant.
Nucleic Acids Res., 53, 2025
|
|
8XXG
 
 | | Crystal structure of human 8-oxoguanine glycosylase K249H mutant bound to the reaction intermediate derived from the crystal soaked into the solution at pH 4.0 under 277 K for 2.5 hours | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*CP*A)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*TP*AP*CP*C)-3'), ... | | Authors: | Unno, M, Koga, M, Minowa, N, Tanaka, Y. | | Deposit date: | 2024-01-18 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Capturing a glycosylase reaction intermediate in DNA repair by freeze-trapping of a pH-responsive hOGG1 mutant.
Nucleic Acids Res., 53, 2025
|
|
8R14
 
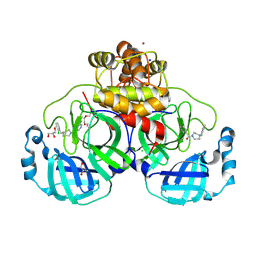 | | Structure of compound 11 bound to SARS-CoV-2 main protease | | Descriptor: | (5-chloranylpyridin-3-yl)-[4-[(2-chlorophenyl)methyl]-1,4-diazepan-1-yl]methanone, 3C-like proteinase, BROMIDE ION, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.336 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches.
Rsc Med Chem, 15, 2024
|
|
8XXK
 
 | | Crystal structure of human 8-oxoguanine glycosylase K249H mutant bound to the reaction intermediate derived from the crystal soaked into the solution at pH 4.0 under 298 K for 3 weeks | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*CP*A)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*TP*AP*CP*C)-3'), ... | | Authors: | Unno, M, Koga, M, Minowa, N, Komuro, S, Tanaka, Y. | | Deposit date: | 2024-01-18 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Capturing a glycosylase reaction intermediate in DNA repair by freeze-trapping of a pH-responsive hOGG1 mutant.
Nucleic Acids Res., 53, 2025
|
|
8JYC
 
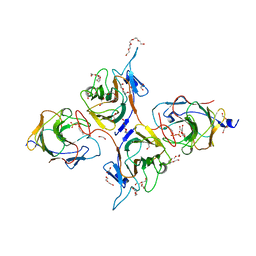 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with DMAPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, Butyrophilin subfamily 3 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
5MM9
 
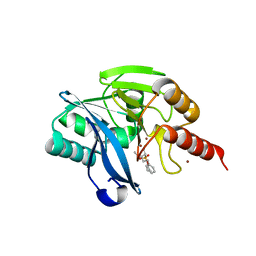 | | VIM-2_2b. Metallo-beta-Lactamase Inhibitors by Bioisosteric Replacement: Preparation, Activity and Binding | | Descriptor: | (2~{R})-2-diethoxyphosphoryl-5-phenyl-pentane-1-thiol, MAGNESIUM ION, Metallo-beta-lactamase VIM-17, ... | | Authors: | Skagseth, S, Akhter, S, Paulsen, M.H, Samuelsen, O, Muhammad, Z, Leiros, H.-K.S, Bayer, A. | | Deposit date: | 2016-12-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metallo-beta-lactamase inhibitors by bioisosteric replacement: Preparation, activity and binding.
Eur J Med Chem, 135, 2017
|
|
8K3W
 
 | | S. cerevisiae Chs1 in complex with UDP-GlcNAc and GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1, MAGNESIUM ION, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8AAD
 
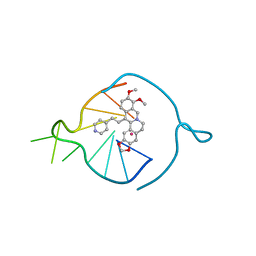 | | Two carbons pendant pyridine derivative of the natural alkaloid Berberine as Human Telomeric G-quadruplex Binder | | Descriptor: | 16,17-dimethoxy-21-(2-pyridin-4-ylethyl)-5,7-dioxa-13$l^{4}-azapentacyclo[11.8.0.0^{2,10}.0^{4,8}.0^{15,20}]henicosa-1(13),2,4(8),9,14,16,18,20-octaene, DNA TAGGGTTAGGGT, POTASSIUM ION | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Papi, F. | | Deposit date: | 2022-07-01 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing the Efficiency of 13-Pyridylalkyl Berberine Derivatives to Human Telomeric G-Quadruplexes Binding: Spectroscopic, Solid State and In Silico Analysis.
Int J Mol Sci, 23, 2022
|
|
7AJ9
 
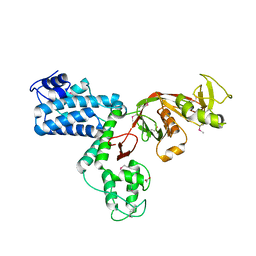 | |
5JPE
 
 | | Yeast-specific serine/threonine protein phosphatase (PPZ1) of Candida albicans | | Descriptor: | CITRATE ANION, GLYCEROL, Serine/threonine-protein phosphatase | | Authors: | Choy, M.S, Chen, E.H, Peti, W, Page, R. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Molecular Insights into the Fungus-Specific Serine/Threonine Protein Phosphatase Z1 in Candida albicans.
Mbio, 7, 2016
|
|
8F4B
 
 | |
7ZUR
 
 | | Four carbons pendant pyridine derivative of the natural alkaloid Berberine as Human Telomeric G-quadruplex Binder | | Descriptor: | 16,17-dimethoxy-21-(4-pyridin-4-ylbutyl)-5,7-dioxa-13$l^{4}-azapentacyclo[11.8.0.0^{2,10}.0^{4,8}.0^{15,20}]henicosa-1(21),2(10),3,8,13,15,17,19-octaene, DNA TAGGGTTAGGGT, POTASSIUM ION | | Authors: | Bazzicalupi, C, Ferraroni, M, Gratteri, P, Papi, F. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Efficiency of 13-Pyridylalkyl Berberine Derivatives to Human Telomeric G-Quadruplexes Binding: Spectroscopic, Solid State and In Silico Analysis.
Int J Mol Sci, 23, 2022
|
|
7ZYK
 
 | | Compound 9 Bound to CK2alpha | | Descriptor: | 2-(5-bromanyl-6-chloranyl-1~{H}-indol-3-yl)ethanenitrile, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
8DB9
 
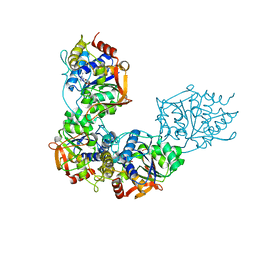 | | Adenosine/guanosine nucleoside hydrolase bound to inhibitor | | Descriptor: | 1-beta-D-ribofuranosyl-1H-1,2,4-triazole-3-carboximidamide, CALCIUM ION, Inosine-uridine preferring nucleoside hydrolase family protein | | Authors: | Muellers, S.N, Allen, K.N, Stockman, B.J. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-Guided Insight into the Specificity and Mechanism of a Parasitic Nucleoside Hydrolase.
Biochemistry, 61, 2022
|
|
6YIK
 
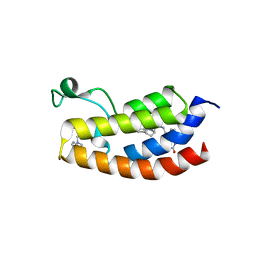 | | Crystal structure of the CREBBP bromodomain in complex with a tetrahydroquinoxaline ligand | | Descriptor: | (3~{R})-~{N}-[3-(3,4-dihydro-2~{H}-quinolin-1-yl)-2,2-bis(fluoranyl)propyl]-3-methyl-2-oxidanylidene-3,4-dihydro-1~{H}-quinoxaline-5-carboxamide, CREBBP | | Authors: | Picaud, S, Brand, M, Tobias, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Conway, S, Filippakopoulos, P. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the CREBBP bromodomain in complex with a tetrahydroquinoxaline ligand
To Be Published
|
|
6VL4
 
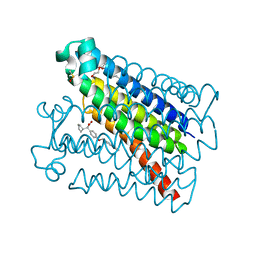 | | Crystal Structure of mPGES-1 bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, Prostaglandin E synthase, TETRAETHYLENE GLYCOL, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6PO4
 
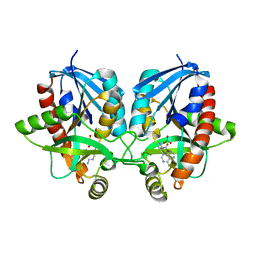 | | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII. | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII.
To Be Published
|
|
9NIQ
 
 | |
8K3Q
 
 | | S. cerevisiae Chs1 in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1 | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8K3V
 
 | | S. cerevisiae Chs1 in complex with UDP-GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1, MAGNESIUM ION, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8DB8
 
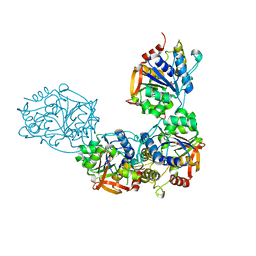 | | Adenosine/guanosine nucleoside hydrolase bound to ImH | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, Inosine-uridine preferring nucleoside hydrolase family protein | | Authors: | Muellers, S.N, Allen, K.N, Stockman, B.J. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-Guided Insight into the Specificity and Mechanism of a Parasitic Nucleoside Hydrolase.
Biochemistry, 61, 2022
|
|
8K3T
 
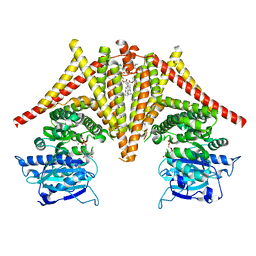 | | S. cerevisiae Chs1 in complex with UDP | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1, MAGNESIUM ION, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
