7S0B
 
 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody N-612-056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-612-056 Fab Heavy Chain, N-612-056 Light Chain, ... | | Authors: | Tanaka, S, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rapid identification of neutralizing antibodies against SARS-CoV-2 variants by mRNA display.
Cell Rep, 38, 2022
|
|
5CSH
 
 | | Crystal Structure of CK2alpha with Compound 4 bound | | Descriptor: | 1-(2-chlorobiphenyl-4-yl)methanamine, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
4UZ5
 
 | |
7S2H
 
 | | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation) | | Descriptor: | CITRATE ANION, Glucokinase 1, putative, ... | | Authors: | Kearns, S.P, Daneshian, L, Swartz, P.D, Carey, S.M, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation)
To Be Published
|
|
6YYI
 
 | | Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase, CITRIC ACID, ... | | Authors: | Lafite, P, Daniellou, R, Bretagne, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of Dictyoglomus thermophilum beta-d-xylosidase DtXyl unravels the structural determinants for efficient notoginsenoside R1 hydrolysis.
Biochimie, 181, 2020
|
|
7RL2
 
 | |
5CTD
 
 | | Crystal structure of the type IX collagen NC2 hetero-trimerization domain with a guest fragment a2a1a1 of type I collagen | | Descriptor: | Collagen alpha-1(I) chain,Collagen alpha-1(IX) chain, Collagen alpha-1(I) chain,Collagen alpha-3(IX) chain, Collagen alpha-2(I) chain,Collagen alpha-2(IX) chain | | Authors: | Boudko, S.P, Bachinger, H.P. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5991 Å) | | Cite: | Structural insight for chain selection and stagger control in collagen.
Sci Rep, 6, 2016
|
|
8RKR
 
 | | Structure of human DELTA-1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE (ALDH4A1) complexed with a monophosphate-tweezer | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-22-(Dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaen-3-ol, BENZOIC ACID, Delta-1-pyrroline-5-carboxylate dehydrogenase, ... | | Authors: | Porfetye, A.T, Vetter, I.R. | | Deposit date: | 2023-12-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | How Do Molecular Tweezers Bind to Proteins? Lessons from X-ray Crystallography.
Molecules, 29, 2024
|
|
4WDH
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation Y168A | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ACETIC ACID, CHLORIDE ION | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
5XQJ
 
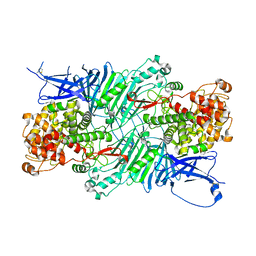 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-[beta-D-galactopyranose-(1-4)]alpha-L-rhamnopyranose, 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
7RR0
 
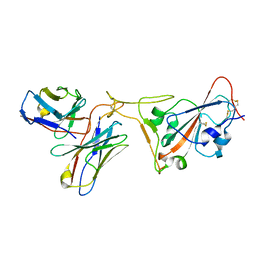 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 222 Fab Heavy Chain, PDI 222 Fab Light Chain, ... | | Authors: | Pymm, P, Glukhova, A, Black, K.A, Tham, W.H. | | Deposit date: | 2021-08-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
5CZY
 
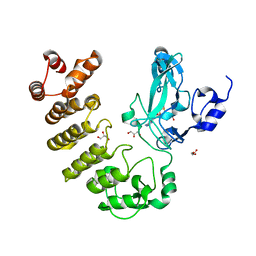 | | Crystal structure of LegAS4 | | Descriptor: | GLYCEROL, Legionella effector LegAS4, S-ADENOSYLMETHIONINE | | Authors: | Son, J, Hwang, K.Y, Lee, W.C. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Legionella pneumophila type IV secretion system effector LegAS4
Biochem.Biophys.Res.Commun., 465, 2015
|
|
4WE1
 
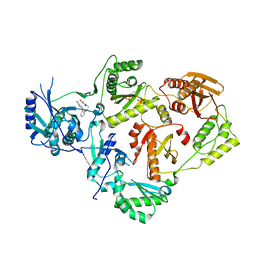 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ600) | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}naphthalene-2-carbonitrile, Gag-Pol polyprotein, MAGNESIUM ION, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Picomolar Inhibitors of HIV-1 Reverse Transcriptase: Design and Crystallography of Naphthyl Phenyl Ethers.
Acs Med.Chem.Lett., 5, 2014
|
|
6YZ5
 
 | | H11-D4 complex with SARS-CoV-2 RBD | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Naismith, J.H, Huo, J, Mikolajek, H, Ward, P, Dumoux, M, Owens, R.J, Le Bas, A. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | H11-D4 complex with SARS-CoV-2 RBD
To Be Published
|
|
4WED
 
 | | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti | | Descriptor: | ABC transporter, periplasmic solute-binding protein, FORMIC ACID, ... | | Authors: | Shabalin, I.G, Otwinowski, Z, Bacal, P, Cymborowski, M.T, Handing, K.B, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti
to be published
|
|
5CTP
 
 | | Crystal structure of CK2alpha with N-(3-(3-chloro-4-(phenyl)benzylamino)propyl)acetamide bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, N-(3-{[(2-chlorobiphenyl-4-yl)methyl]amino}propyl)acetamide, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
7S2N
 
 | |
5D13
 
 | |
7SBC
 
 | |
4V27
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, ... | | Authors: | Hakki, Z, Bellmaine, S, Thompson, A.J, Speciale, G, Davies, G.J, Williams, S.J. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Kinetic Dissection of the Endo-Alpha-1,2-Mannanase Activity of Bacterial Gh99 Glycoside Hydrolases from Bacteroides Spp.
Chemistry, 21, 2015
|
|
7RK2
 
 | | Crystal structure of the human astrovirus serotype 8 capsid spike in complex with scFv 2D9, an astrovirus-neutralizing antibody, at 2.65-A resolution | | Descriptor: | Capsid protein VP25, scFv 2D9 | | Authors: | Meyer, L, Cuellar, C, DuBois, R.M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Two Human Astrovirus Capsid/Neutralizing Antibody Complexes Reveal Distinct Epitopes and Inhibition of Virus Attachment to Cells.
J.Virol., 96, 2022
|
|
8RBV
 
 | | SARS-CoV-2 Spike-derived peptide S976-984 S982A mutant (VLNDILARL) presented by HLA-A*02:01 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
7S2P
 
 | | Crystal structure of the F337L mutation of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN | | Descriptor: | 2-{[(benzyloxy)carbonyl]amino}-2-deoxy-beta-D-glucopyranose, Glucokinase 1, SULFATE ION | | Authors: | Carey, S.M, Nettles, R.B, Daneshian, L, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the F337L mutation of Trypanosoma cruzi glucokinase in complex with inhibitor CBZ-GlcN
To Be Published
|
|
5XUG
 
 | | Complex structure(RmMan134A-M5). | | Descriptor: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, endo-1,4-beta-mannanase | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-06-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5CW4
 
 | | Structure of CfBRCC36-CfKIAA0157 complex (Selenium Edge) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, GLYCEROL, Protein FAM175B, ... | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
