7OKA
 
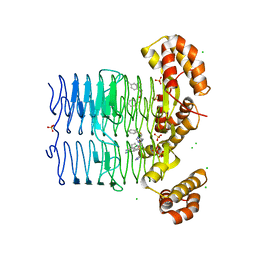 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 14 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-(1~{H}-imidazol-4-ylmethyl)ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, ... | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OJQ
 
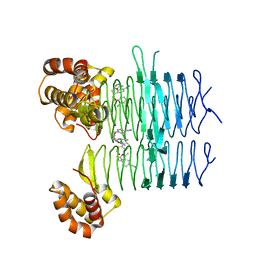 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 7 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-[(5-methyl-1,3,4-oxadiazol-2-yl)methyl]ethanamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
8Z4S
 
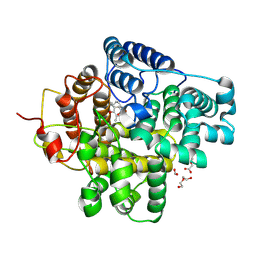 | | The crystal structure of a Hydroquinone Dioxygenase PaD with nonnatural substrate S6 | | Descriptor: | 2,3,5-trimethylbenzene-1,4-diol, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Z.W, Huang, J.-W, Wang, Y.X, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-04-17 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Substrate specificity of a branch of aromatic dioxygenases determined by three distinct motifs.
Nat Commun, 15, 2024
|
|
7OJW
 
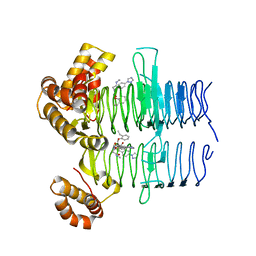 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 93 | | Descriptor: | 2-[2-(2-ethylphenoxy)ethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-~{N}-methyl-ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7NX1
 
 | | TG domain of LTK | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Leukocyte tyrosine kinase receptor, TERBIUM(III) ION | | Authors: | De Munck, S, Savvides, S.N. | | Deposit date: | 2021-03-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of cytokine-mediated activation of ALK family receptors.
Nature, 600, 2021
|
|
8KI6
 
 | | Structure of tomato spotted wilt virus L protein binding to Ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, RNA-directed RNA polymerase L | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin.
Nat.Plants, 11, 2025
|
|
6UZF
 
 | | Crystal structure of the unliganded bromodomain of human BRD9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, DIMETHYL SULFOXIDE, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
1EPR
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-135,040 | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Badasso, M, Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
8K4K
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Olsalazine carbon derivative | | Descriptor: | 5-[(E)-2-(3-carboxy-4-oxidanyl-phenyl)ethenyl]-2-oxidanyl-benzoic acid, Flavin reductase (NADPH), GLYCEROL, ... | | Authors: | Ha, J.H, Jung, H.M, dela Cerna, M.V.C, Burlison, J.A, Lee, D, Ryu, K.S. | | Deposit date: | 2023-07-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Innovative Inhibitor with a New Chemical Moiety Aimed at Biliverdin IX beta Reductase for Thrombocytopenia and Resilient against Cellular Degradation.
Pharmaceutics, 16, 2024
|
|
8Z9F
 
 | | Crystal structure of glyoxylate reductase from Acetobacter aceti in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-hydroxyisobutyrate dehydrogenase | | Authors: | Majumder, T.R, Yoshizawa, T, Inoue, M, Aono, R, Matsumura, H, Mihara, H. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the mechanism underlying the dual cofactor specificity of glyoxylate reductase from Acetobacter aceti in the beta-hydroxyacid dehydrogenase family.
Biochim Biophys Acta Proteins Proteom, 1873, 2025
|
|
7OKK
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 12e | | Descriptor: | 1,2-ETHANEDIOL, 2-[[6-[(2-chloranyl-3-cyano-pyridin-4-yl)amino]-2-oxidanylidene-1H-quinolin-4-yl]amino]-N-methyl-ethanamide, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
8EKS
 
 | |
8Z2G
 
 | | MHET bound form of PET-degrading cutinase mutant Cut190*SS_S176A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, AMMONIUM ION, ... | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8EM3
 
 | |
6VDX
 
 | | Crystal Structure of Dehaloperoxidase B in Complex with cofactor Manganese(III)- Porphyrin and Substrate Trichlorophenol | | Descriptor: | 1,2-ETHANEDIOL, 2,4,6-trichlorophenol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, McGuire, A, Malewschik, T. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Nonnative Heme Incorporation into Multifunctional Globin Increases Peroxygenase Activity an Order and Magnitude Compared to Native Enzyme
To Be Published
|
|
8OVS
 
 | |
6VDU
 
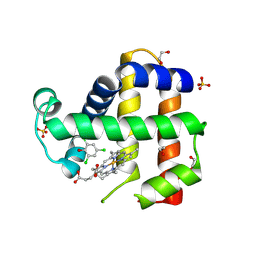 | | Crystal Structure of Dehaloperoxidase B in Complex with cofactor Iron(III) Deuteroporphyrin IX and Substrate Trichlorophenol | | Descriptor: | 1,2-ETHANEDIOL, 2,4,6-trichlorophenol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, McGuire, A, Malewschik, T. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Nonnative Heme Incorporation into Multifunctional Globin Increases Peroxygenase Activity an Order and Magnitude Compared to Native Enzyme
To Be Published
|
|
8ONO
 
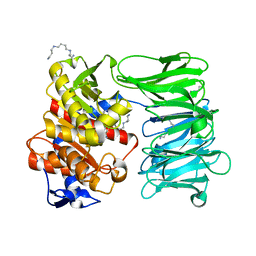 | | Modified oligopeptidase B from S. proteamaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2023-04-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution
To Be Published
|
|
6VB4
 
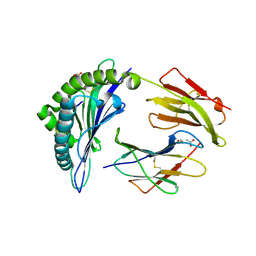 | | HLA-B*15:02 complexed with a synthetic peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Schutte, R.J, Li, D, Andring, J, McKenna, R, Ostrov, D.A. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | HLA-B*15:02 complexed with a synthetic peptide
To Be Published
|
|
6VH1
 
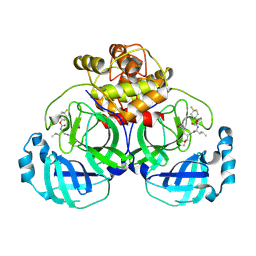 | | 2.30 A resolution structure of MERS 3CL protease in complex with inhibitor 6h | | Descriptor: | N~2~-{[(4,4-difluorocyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
8F1Z
 
 | | EGFR kinase in complex with Bayer #33 | | Descriptor: | (2P)-3-(3-chloro-2-methoxyanilino)-2-[3-(2-methoxy-2-methylpropoxy)pyridin-4-yl]-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, CITRIC ACID, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OWG
 
 | | Crystal structure of D1228V c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8F1X
 
 | | EGFR kinase in complex with mobocertinib (TAK-788) | | Descriptor: | Epidermal growth factor receptor, propan-2-yl 2-[[4-[2-(dimethylamino)ethyl-methyl-amino]-2-methoxy-5-(propanoylamino)phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-11-06 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical analysis of EGFR exon20 insertion variants insASV and insSVD and their inhibitor sensitivity.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8F5X
 
 | | Crystal structure of human eosinophil-derived neurotoxin (EDN, ribonuclease 2) in complex with 5'-adenosine monophosphate (AMP) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Non-secretory ribonuclease, ... | | Authors: | Tran, T.T.Q, Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ancestral sequence reconstruction dissects structural and functional differences among eosinophil ribonucleases.
J.Biol.Chem., 300, 2024
|
|
8OF7
 
 | | Cyc15 Diels Alderase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Back, C.R, Barringer, R.W.L, Zorn, K, Manzo-Ruiz, M, Race, P.R. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Interrogation of an Enzyme Library Reveals the Catalytic Plasticity of Naturally Evolved [4+2] Cyclases.
Chembiochem, 24, 2023
|
|
