5O1Y
 
 | | Structure of Nrd1 RNA binding domain in complex with RNA (GUAA) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, ... | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
1U4B
 
 | | Extension of an adenine-8oxoguanine mismatch | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
4RZR
 
 | | Bypass of a bulky adduct dG1,8 by DPO4 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Vyas, R, Suo, Z. | | Deposit date: | 2014-12-23 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Basis for the Bypass of a Bulky DNA Adduct Catalyzed by a Y-Family DNA Polymerase.
J.Am.Chem.Soc., 137, 2015
|
|
3LP3
 
 | | p15 HIV RNaseH domain with inhibitor MK3 | | Descriptor: | 3-[4-(diethylamino)phenoxy]-6-(ethoxycarbonyl)-5,8-dihydroxy-7-oxo-7,8-dihydro-1,8-naphthyridin-1-ium, MANGANESE (II) ION, p15 | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
4BWM
 
 | | KlenTaq mutant in complex with a RNA/DNA hybrid | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOCP)-3', 5'-R(*AP*AP*AP*GP*GP*GP*CP*GP*CP*CP*GP*UP*GP*GP*UP*C)-3', ... | | Authors: | Blatter, N, Bergen, K, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2013-07-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure and Function of an RNA-Reading Thermostable DNA Polymerase.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4OL8
 
 | | Ty3 reverse transcriptase bound to DNA/RNA | | Descriptor: | 5'-D(*CP*AP*TP*CP*TP*TP*CP*CP*TP*CP*TP*CP*TP*CP*TP*C)-3', 5'-R(*CP*UP*GP*AP*GP*AP*GP*AP*GP*AP*GP*GP*AP*AP*GP*AP*UP*G)-3', Reverse transcriptase/ribonuclease H, ... | | Authors: | Nowak, E, Miller, J.T, Bona, M.K, Studnicka, J, Szczepanowski, R.H, Jurkowski, J, Le Grice, S.F.J, Nowotny, M. | | Deposit date: | 2014-01-23 | | Release date: | 2014-03-05 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ty3 reverse transcriptase complexed with an RNA-DNA hybrid shows structural and functional asymmetry.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7DBN
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V/F160M:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
7DBM
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
1U48
 
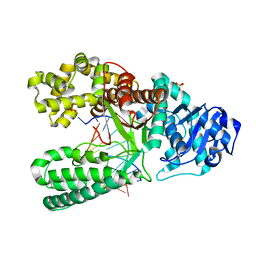 | | Extension of a cytosine-8-oxoguanine base pair | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
5WU4
 
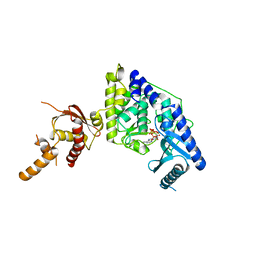 | |
5WU1
 
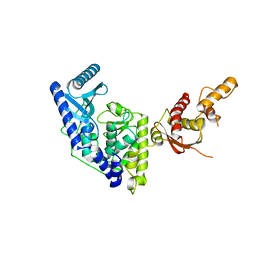 | | Crystal structure of apo human Tut1, form I | | Descriptor: | CHLORIDE ION, Speckle targeted PIP5K1A-regulated poly(A) polymerase | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2016-12-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nat Commun, 8, 2017
|
|
6EPC
 
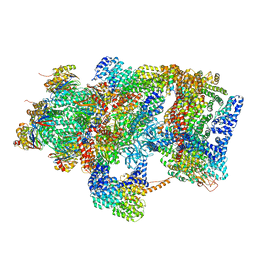 | | Ground state 26S proteasome (GS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPD
 
 | | Substrate processing state 26S proteasome (SPS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPE
 
 | | Substrate processing state 26S proteasome (SPS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPF
 
 | | Ground state 26S proteasome (GS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
5WU5
 
 | | Crystal structure of apo human Tut1, form III | | Descriptor: | Speckle targeted PIP5K1A-regulated poly(A) polymerase | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2016-12-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nat Commun, 8, 2017
|
|
6HY0
 
 | | Atomic models of P1, P4 C-terminal fragment and P8 fitted in the bacteriophage phi6 nucleocapsid reconstructed with icosahedral symmetry | | Descriptor: | Major Outer Capsid Protein P8, Major inner protein P1, Packaging Enzyme P4 | | Authors: | El Omari, K, Ilca, S.L, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2018-10-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus.
Nature, 570, 2019
|
|
5WU2
 
 | |
5WU3
 
 | | Crystal structure of human Tut1 bound with MgUTP, form II | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2016-12-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nat Commun, 8, 2017
|
|
6VAG
 
 | | Crystal structure of the oligomerization domain of phosphoprotein from parainfluenza virus 5 | | Descriptor: | GLYCEROL, Phosphoprotein | | Authors: | Aggarwal, M, Abdella, R, He, Y, Lamb, R.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3IG1
 
 | | HIV-1 Reverse Transcriptase with the Inhibitor beta-Thujaplicinol Bound at the RNase H Active Site | | Descriptor: | 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, HIV-1 Reverse Transcriptase p51 subunit, HIV-1 Reverse Transcriptase p66 subunit, ... | | Authors: | Himmel, D.M, Maegley, K.A, Pauly, T.A, Arnold, E. | | Deposit date: | 2009-07-27 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site.
Structure, 17, 2009
|
|
6AF0
 
 | | Structure of Ctr9, Paf1 and Cdc73 ternary complex from Myceliophthora thermophila | | Descriptor: | Cdc73 protein, Ctr9 protein, Paf1 protein | | Authors: | Wang, Z, Deng, P, Zhou, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W4F
 
 | | Importin binding to pol Mu NLS peptide | | Descriptor: | DNA-directed DNA/RNA polymerase mu, GLYCEROL, Importin subunit alpha-1 | | Authors: | Pedersen, L.C, London, R.E. | | Deposit date: | 2017-06-10 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Variations in nuclear localization strategies among pol X family enzymes.
Traffic, 2018
|
|
3QIP
 
 | | Structure of HIV-1 reverse transcriptase in complex with an RNase H inhibitor and nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5,6-dihydroxy-2-[(2-phenyl-1H-indol-3-yl)methyl]pyrimidine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Lansdon, E.B, Kirschberg, T.A. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0926 Å) | | Cite: | Structural and Binding Analysis of Pyrimidinol Carboxylic Acid and N-Hydroxy Quinazolinedione HIV-1 RNase H Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
4UXB
 
 | | Human ARTD1 (PARP1) - Catalytic domain in complex with inhibitor PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, POLY ADP-RIBOSE POLYMERASE 1, SULFATE ION | | Authors: | Tresaugues, L, Thorsell, A.G, Karlberg, T, Schuler, H. | | Deposit date: | 2014-08-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
