6BTW
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a phenyl-pteridinone inhibitor | | Descriptor: | 2-[(3,5-difluoro-4-hydroxyphenyl)amino]-8-phenyl-7,8-dihydropteridin-6(5H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-07 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a phenyl-pteridinone inhibitor
To Be Published
|
|
6WW3
 
 | | Crystal structure of HERC2 ZZ domain in complex with SUMO1 tail | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SUMO1 linked HERC2 ZZ domain (Small ubiquitin-related modifier 1,E3 ubiquitin-protein ligase HERC2), ... | | Authors: | Liu, J, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1.
Structure, 28, 2020
|
|
6WW8
 
 | | BRD4 Bromodomain 1 in complex with triple CDK4/6-PI3K-BET inhibitor | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-({4-[5-(morpholin-4-yl)-7-oxo-7H-thieno[3,2-b]pyran-3-yl]phenyl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Bromodomain-containing protein 4 | | Authors: | Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-08 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | A triple action CDK4/6-PI3K-BET inhibitor with augmented cancer cell cytotoxicity.
Cell Discov, 6, 2020
|
|
6BOY
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET6 PROTAC. | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(8-{[({2-[(3S)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}oxy)acetyl]amino}octyl)acetamide, Bromodomain-containing protein 4, DNA damage-binding protein 1, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-21 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6X2Y
 
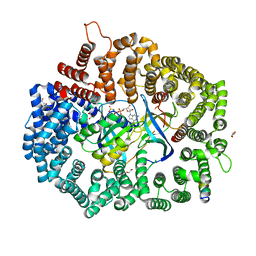 | |
6BQT
 
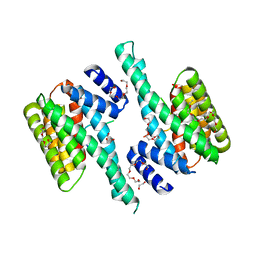 | |
6X4Q
 
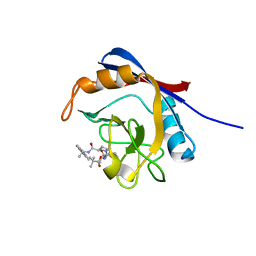 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 33) | | Descriptor: | (2R,5S,11S,14S,18E)-14-cyclobutyl-2,11,17,17-tetramethyl-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
6X2R
 
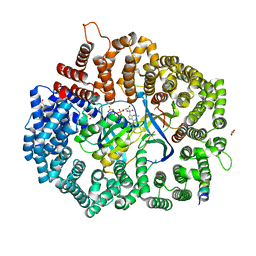 | | Crystal Structure of the 4E-TNES peptide bound to CRM1 | | Descriptor: | Eukaryotic translation initiation factor 4E transporter, Exportin-1, GLYCEROL, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6VUB
 
 | |
6VUF
 
 | |
6VKO
 
 | |
6VPJ
 
 | |
6VVO
 
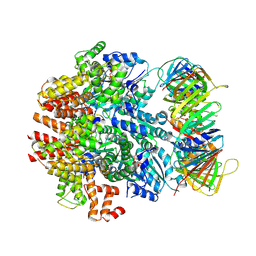 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
6D9X
 
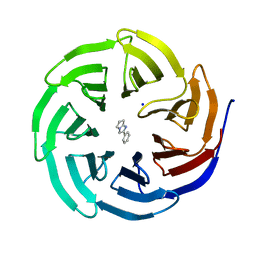 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | 2-phenyl-6,7-dihydro-5H-pyrrolo[1,2-a]imidazole, SODIUM ION, WD repeat-containing protein 5 | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6BU6
 
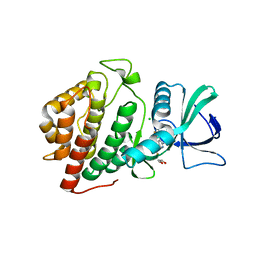 | | Crystal Structure of the Human vaccinia-related kinase bound to a bis-difluorophenol-aminopyridine inhibitor | | Descriptor: | 4,4'-(2-aminopyridine-3,5-diyl)bis(2,6-difluorophenol), CHLORIDE ION, GLYCEROL, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Santiago, A.S, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-08 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a bis-difluorophenol-aminopyridine inhibitor
To Be Published
|
|
6BGV
 
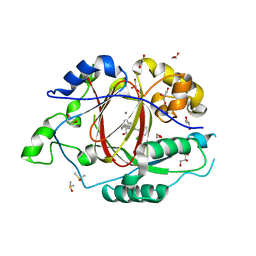 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N40) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH5
 
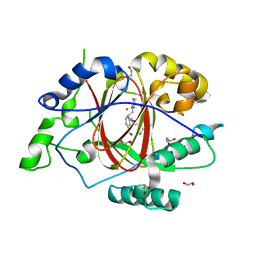 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(3-(piperidin-1-yl)propoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N48) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[3-(piperidin-1-yl)propoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGG
 
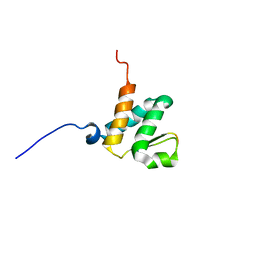 | | Solution NMR structures of the BRD3 ET domain in complex with a CHD4 peptide | | Descriptor: | Bromodomain-containing protein 3, CHD4 | | Authors: | Wai, D.C.C, Szyszka, T.N, Campbell, A.E, Kwong, C, Wilkinson-White, L, Silva, A.P.G, Low, J.K.K, Kwan, A.H, Gamsjaeger, R, Lu, B, Vakoc, C.R, Blobel, G.A, Mackay, J.P. | | Deposit date: | 2017-10-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.
J. Biol. Chem., 293, 2018
|
|
6BN7
 
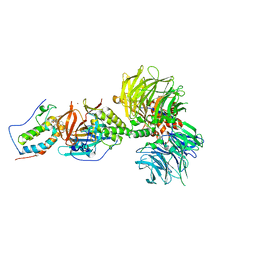 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET23 PROTAC. | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BP0
 
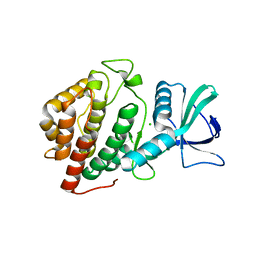 | | Crystal Structure of the Human vaccinia-related kinase 1 bound to (R)-2-phenylaminopteridinone inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7,8-trimethyl-7,8-dihydropteridin-6(5H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase 1 bound to (R)-2-phenylaminopteridinone inhibitor
To Be Published
|
|
6BRU
 
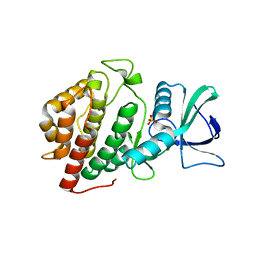 | | Crystal Structure of the Human vaccinia-related kinase bound to a (S)-2-phenylaminopteridinone inhibitor | | Descriptor: | (7S)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7,8-trimethyl-7,8-dihydropteridin-6(5H)-one, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Aquino, B, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a (S)-2-phenylaminopteridinone inhibitor
To Be Published
|
|
6WHO
 
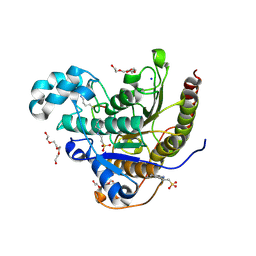 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHN
 
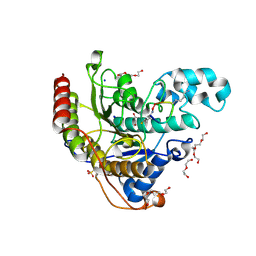 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHQ
 
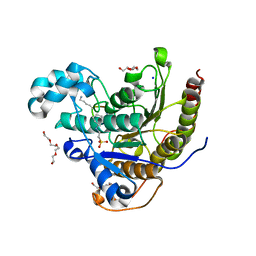 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHZ
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | Histone deacetylase 2, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
