6H03
 
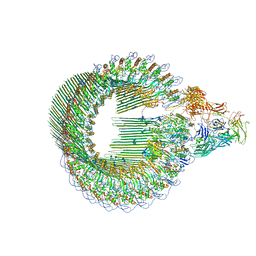 | | OPEN CONFORMATION OF THE MEMBRANE ATTACK COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5,Complement C5, ... | | Authors: | Menny, A, Serna, M, Boyd, C.M, Gardner, S, Joseph, A.P, Topf, M, Bubeck, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | CryoEM reveals how the complement membrane attack complex ruptures lipid bilayers.
Nat Commun, 9, 2018
|
|
6GZK
 
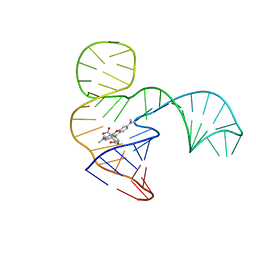 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | Descriptor: | 5-carboxy methylrhodamine, TMR3 (48-MER) | | Authors: | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | Deposit date: | 2018-07-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
6H04
 
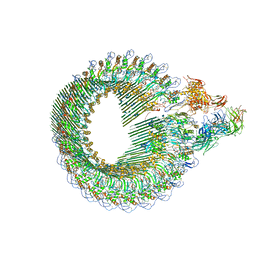 | | Closed conformation of the Membrane Attack Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5,Complement C5, ... | | Authors: | Menny, A, Serna, M, Boyd, C.M, Gardner, S, Joseph, A.P, Topf, M, Bubeck, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | CryoEM reveals how the complement membrane attack complex ruptures lipid bilayers.
Nat Commun, 9, 2018
|
|
5L46
 
 | |
6RF7
 
 | | Crystal structure of the light-driven sodium pump KR2 in the monomeric form, pH 8.9 | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6H48
 
 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | Descriptor: | Orf20 | | Authors: | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
5L8S
 
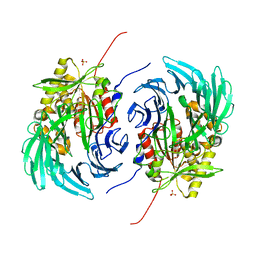 | | The crystal structure of a cold-adapted acylaminoacyl peptidase reveals a novel quaternary architecture based on the arm-exchange mechanism | | Descriptor: | Amino acyl peptidase, SULFATE ION | | Authors: | Brocca, S, Ferrari, C, Barbiroli, A, Pesce, A, Lotti, M, Nardini, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A bacterial acyl aminoacyl peptidase couples flexibility and stability as a result of cold adaptation.
FEBS J., 283, 2016
|
|
6H52
 
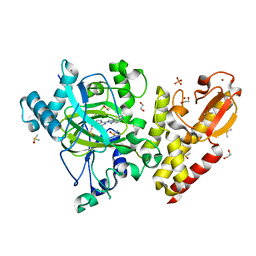 | | Crystal structure of human KDM5B in complex with compound 34g | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-(1-cyclopentylpiperidin-4-yl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6RNX
 
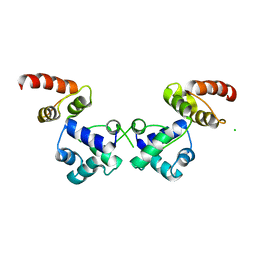 | | Crystal structure of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
2MOX
 
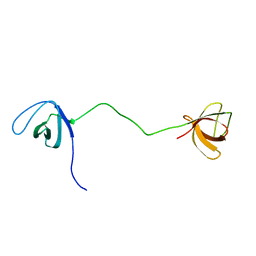 | | solution structure of tandem SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Zhao, D, Wang, C, Zhang, J, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2014-05-07 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
6RQP
 
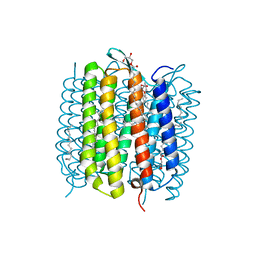 | | Steady-state-SMX dark state structure of bacteriorhodopsin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, A, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
5LT0
 
 | |
5LTA
 
 | |
6RUZ
 
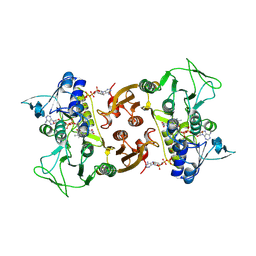 | | NADH-dependent Coenzyme A Disulfide Reductase | | Descriptor: | COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, NADH oxidase | | Authors: | Koepke, J, Preu, J. | | Deposit date: | 2019-05-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization and X-ray structure of the NADH-dependent coenzyme A disulfide reductase from Thermus thermophilus.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
5LHG
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor 4-methyl-N-[4-[[4-(1-methylpiperidin-4-yl)oxyphenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | 4-methyl-N-[4-[[4-[(1-methyl-4-piperidyl)oxy]phenoxy]methyl]phenyl]thieno[3,2-b]pyrrole-5-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-11 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
6DEK
 
 | |
6D87
 
 | |
6RVO
 
 | |
6DER
 
 | | Crystal structure of Candida albicans acetohydroxyacid synthase in complex with the herbicide metosulam | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Guddat, L.W. | | Deposit date: | 2018-05-12 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.126 Å) | | Cite: | Commercial AHAS-inhibiting herbicides are promising drug leads for the treatment of human fungal pathogenic infections.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6RVH
 
 | | NADH-dependent Coenzyme A Disulfide Reductase soaked with Menadione | | Descriptor: | COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, MENADIONE, ... | | Authors: | Koepke, J, Preu, J. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterization and X-ray structure of the NADH-dependent coenzyme A disulfide reductase from Thermus thermophilus.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
6RYV
 
 | | Copper oxidase from Colletotrichum graminicola | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Kelch domain-containing protein, ... | | Authors: | Offen, W.A, Henrissat, B, Davies, G.J. | | Deposit date: | 2019-06-12 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Fungal Copper Radical Oxidase with High Catalytic Efficiency toward 5-Hydroxymethylfurfural and Benzyl Alcohols for Bioprocessing
Acs Catalysis, 2020
|
|
5M28
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to maltotriose | | Descriptor: | CHLORIDE ION, MalE1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2016-10-12 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.079 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
5LCV
 
 | |
6DQ7
 
 | |
5LKY
 
 | | X-ray crystal structure of N-acetylneuraminic acid lyase in complex with pyruvate, with the phenylalanine at position 190 replaced with the non-canonical amino acid dihydroxypropylcysteine. | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-acetylneuraminate lyase | | Authors: | Windle, C.L, Trinh, C.H, Pearson, A.R, Nelson, A.S, Berry, A. | | Deposit date: | 2016-07-25 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending enzyme molecular recognition with an expanded amino acid alphabet.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
