6O9U
 
 | | KirBac3.1 at a resolution of 2 Angstroms | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3,3',3''-phosphoryltripropanoic acid, BARIUM ION, ... | | Authors: | Gulbis, J.M, Clarke, O.B. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
6OBA
 
 | | The beta2 adrenergic receptor bound to a negative allosteric modulator | | Descriptor: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, 6-bromo-N~2~-phenylquinazoline-2,4-diamine, Beta-2 adrenergic receptor,Lysozyme,Beta-2 adrenergic receptor, ... | | Authors: | Liu, X, Stobel, A, Kaindl, J, Dengler, D, ClarK, M, Mahoney, J, Korczynska, M, Matt, R.A, Hubner, H, Xu, X, Stanek, M, Hirata, K, Shoichet, B, Sunahara, R, Gmeiner, R, Kobilka, B.K. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An allosteric modulator binds to a conformational hub in the beta2adrenergic receptor.
Nat.Chem.Biol., 16, 2020
|
|
7OTC
 
 | | Cryo-EM structure of an Escherichia coli 70S ribosome in complex with elongation factor G and the antibiotic Argyrin B | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Wieland, M, Koller, T.O, Wilson, D.N. | | Deposit date: | 2021-06-10 | | Release date: | 2022-05-11 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cyclic octapeptide antibiotic argyrin B inhibits translation by trapping EF-G on the ribosome during translocation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RL2
 
 | |
8YZW
 
 | |
8YZZ
 
 | | The structure of HLA-A*2402 complex with peptide from SARS-CoV-2 S448-456 NYNYLYRLF(Prototype) | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Spike protein S1 | | Authors: | Shang, B.L, Zhang, J.N, Tian, J.M, Liu, J. | | Deposit date: | 2024-04-08 | | Release date: | 2025-01-29 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | T cell immune evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots.
Nat.Immunol., 26, 2025
|
|
6OS0
 
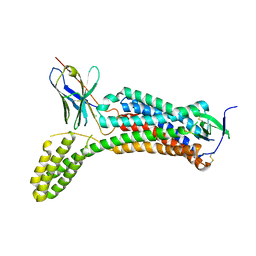 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to angiotensin II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensinogen, CHLORIDE ION, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
8RG4
 
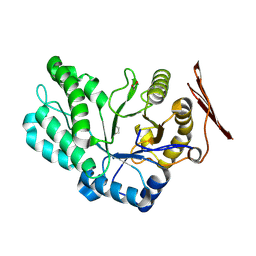 | | Crystal structure of PbFucA from Planctomycetes bacterium K23_9 in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, Glycoside-hydrolase family GH114 TIM-barrel domain-containing protein | | Authors: | Perez-Cruz, C, Moraleda-Montoya, A, Liebana, R, Lorizate, M, Arrizabalaga, U, Garcia-Alija, M, Terrones, O, Contreras, F.X, Guerin, M.E, Trastoy, B, Alonso-Saez, L. | | Deposit date: | 2023-12-13 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Mechanisms of recalcitrant fucoidan breakdown in marine Planctomycetota.
Nat Commun, 15, 2024
|
|
7PH3
 
 | | AMP-PNP bound nanodisc reconstituted MsbA with nanobodies, spin-labeled at position A60C | | Descriptor: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent lipid A-core flippase, ... | | Authors: | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
9CLD
 
 | | Crystal structure of maltose binding protein (Apo) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Habel, E, Frkic, R.L, Jackson, C.J, Huber, T, Otting, G. | | Deposit date: | 2024-07-10 | | Release date: | 2024-12-18 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Rendering Proteins Fluorescent Inconspicuously: Genetically Encoded 4-Cyanotryptophan Conserves Their Structure and Enables the Detection of Ligand Binding Sites.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8Z4W
 
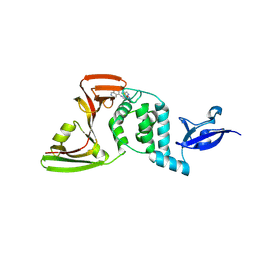 | | Crystal structures of SARS-CoV-2 papain-like protease in complex with covalent inhibitors | | Descriptor: | 1-[4-[[[4-(isoquinolin-5-ylamino)-6-(methylamino)-1,3,5-triazin-2-yl]amino]methyl]piperidin-1-yl]ethanone, GLYCEROL, Non-structural protein 3, ... | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Covalent DNA-Encoded Library Workflow Drives Discovery of SARS-CoV-2 Nonstructural Protein Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
8WJC
 
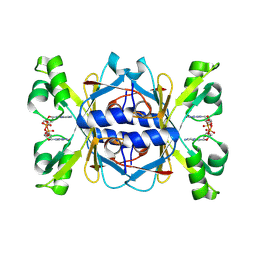 | | Structure of AcrIIA7 complexed with 3'3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-25 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
9K1N
 
 | |
6OUG
 
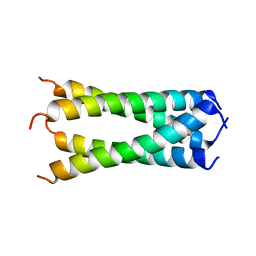 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor, TM + cytosolic helix construct | | Descriptor: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, Matrix protein 2 | | Authors: | Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2019-05-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
8U16
 
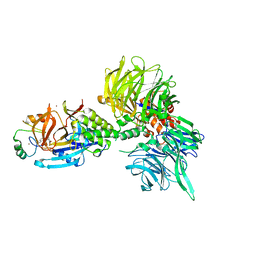 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-short bound to Pomalidomide | | Descriptor: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8WPN
 
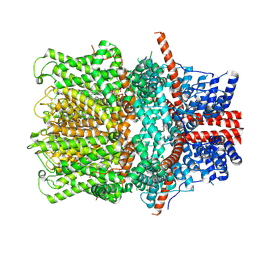 | | Cryo-EM structure of the human TRPC4 in lipid nanodiscs | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM structure of the heteromeric TRPC1/TRPC4 channel.
Nat.Struct.Mol.Biol., 32, 2025
|
|
5XCY
 
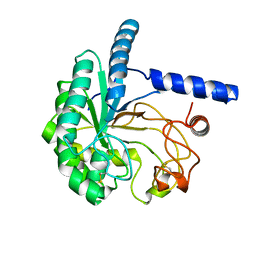 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium at 1.2 angstrom | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6QQN
 
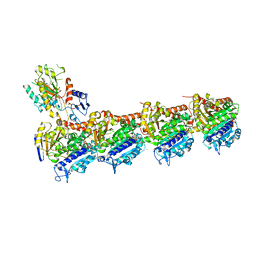 | | Tubulin-TH588 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Patterson, J.C, Joughin, B.A, Prota, A.E, Muehlethaler, T, Jonas, O.H, Whitman, M.A, Varmeh, S, Chen, S, Balk, S.P, Steinmetz, M.O, Lauffenburger, D.A, Yaffe, M.B. | | Deposit date: | 2019-02-18 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | VISAGE Reveals a Targetable Mitotic Spindle Vulnerability in Cancer Cells.
Cell Syst, 9, 2019
|
|
7P43
 
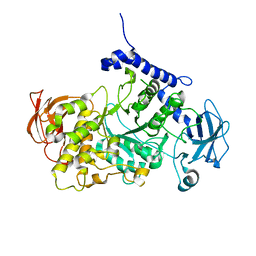 | | Structure of CgGBE in complex with maltotriose | | Descriptor: | 1,4-alpha-glucan-branching enzyme, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
5HPI
 
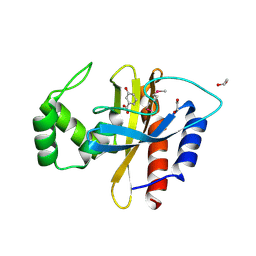 | | Crystal Structure of the Double Mutant of PobR Transcription Factor Inducer Binding Domain-3-Hydroxy Benzoic Acid complex from Acinetobacter | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, SULFATE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrejczak, R, Jha, R, Strauss, C.E.M, Joachimiak, A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-09-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor.
Nucleic Acids Res., 44, 2016
|
|
5XSA
 
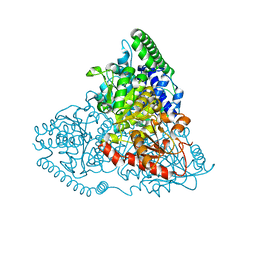 | | Crystal Structure of Transketolase in complex with TPP intermediate III from Pichia Stipitis | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, Transketolase | | Authors: | Li, T.L, Hsu, N.S, Wang, Y.L. | | Deposit date: | 2017-06-13 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.975 Å) | | Cite: | The Mesomeric Effect of Thiazolium on non-Kekule Diradicals in Pichia stipitis Transketolase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5HXQ
 
 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M, E75A and H103Y Mutants) Complexed with DMSPP | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ... | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of a Z,Z-Farnesyl Diphosphate Synthase
Acs Omega, 2, 2017
|
|
6NTH
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by (S) Stereoisomer of A-232 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.421 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
5HXT
 
 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M, E75A and H103Y Mutants) Complexed with IPP and DMSPP | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ... | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of a Z,Z-Farnesyl Diphosphate Synthase
Acs Omega, 2, 2017
|
|
7F44
 
 | | Crystal structure of Moraxella catarrhalis enoyl-ACP-reductase (FabI) in complex with the cofactor NAD | | Descriptor: | CALCIUM ION, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Katiki, M, Neetu, N, Pratap, S, Kumar, P. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Biochemical and structural basis for Moraxella catarrhalis enoyl-acyl carrier protein reductase (FabI) inhibition by triclosan and estradiol.
Biochimie, 198, 2022
|
|
