5IGS
 
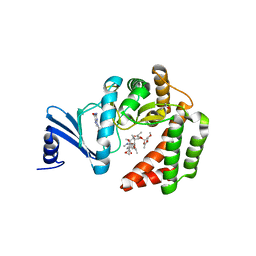 | | Macrolide 2'-phosphotransferase type I - complex with guanosine and oleandomycin | | Descriptor: | (3S,5R,6S,7R,8R,11R,12S,13R,14S,15S)-6-HYDROXY-5,7,8,11,13,15-HEXAMETHYL-4,10-DIOXO-14-{[3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSYL]OXY}-1,9-DIOXASPIRO[2.13]HEXADEC-12-YL 2,6-DIDEOXY-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSIDE, GUANOSINE, ISOPROPYL ALCOHOL, ... | | Authors: | Berghuis, A.M, Fong, D.H. | | Deposit date: | 2016-02-28 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural Basis for Kinase-Mediated Macrolide Antibiotic Resistance.
Structure, 25, 2017
|
|
3B2U
 
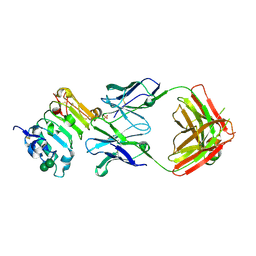 | | Crystal structure of isolated domain III of the extracellular region of the epidermal growth factor receptor in complex with the Fab fragment of IMC-11F8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Ferguson, K.M, Li, S, Kussie, P. | | Deposit date: | 2007-10-19 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis for EGF receptor inhibition by the therapeutic antibody IMC-11F8.
Structure, 16, 2008
|
|
5IWU
 
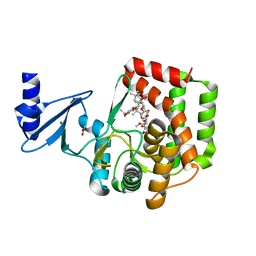 | |
3B2V
 
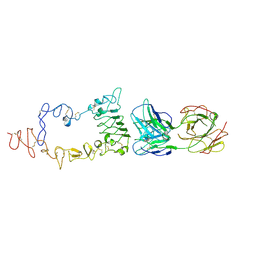 | | Crystal structure of the extracellular region of the epidermal growth factor receptor in complex with the Fab fragment of IMC-11F8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Ferguson, K.M, Li, S, Kussie, P. | | Deposit date: | 2007-10-19 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for EGF receptor inhibition by the therapeutic antibody IMC-11F8.
Structure, 16, 2008
|
|
1G35
 
 | | CRYSTAL STRUCTURE OF HIV-1 PROTEASE IN COMPLEX WITH INHIBITOR, AHA024 | | Descriptor: | 2-[4-(HYDROXY-METHOXY-METHYL)-BENZYL]-7-(4-HYDROXYMETHYL-BENZYL)-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-1LAMBDA6-[1,2,7]THIADIAZEPANE-4,5-DIOL, HIV-1 PROTEASE | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors.
J.Med.Chem., 44, 2001
|
|
1G2K
 
 | | HIV-1 PROTEASE WITH CYCLIC SULFAMIDE INHIBITOR, AHA047 | | Descriptor: | 3-(7-BENZYL-4,5-DIHYDROXY-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-1L6-[1,2,7]THIADIAZEPAN-2-YLMETHYL)-N-METHYL-BENZAMIDE, PROTEASE RETROPEPSIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2000-10-20 | | Release date: | 2001-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors.
J.Med.Chem., 44, 2001
|
|
7ZDQ
 
 | | Cryo-EM structure of Human ACE2 bound to a high-affinity SARS CoV-2 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Bate, N, Savva, C.G, Moody, P.C.E, Brown, E.A, Schwabe, W.R, Brindle, N.P.J, Ball, J.K, Sale, J.E. | | Deposit date: | 2022-03-29 | | Release date: | 2022-05-18 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In vitro evolution predicts emerging SARS-CoV-2 mutations with high affinity for ACE2 and cross-species binding.
Plos Pathog., 18, 2022
|
|
6N4P
 
 | |
1H4P
 
 | | Crystal structure of exo-1,3-beta glucanse from Saccharomyces cerevisiae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUCAN 1,3-BETA-GLUCOSIDASE I/II, GLYCEROL, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2001-05-11 | | Release date: | 2003-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Er Protein Folding Sensor Udp-Glucose Glycoprotein:Glucosyltransferase Modifies Substrates Distant to Local Changes in Glycoprotein Conformation.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4O30
 
 | | Crystal structure of ATXR5 in complex with histone H3.1 and AdoHcy | | Descriptor: | BETA-MERCAPTOETHANOL, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase ATXR6, ... | | Authors: | Bergamin, E, Mongeon, V, Couture, J.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication.
Science, 343, 2014
|
|
7B4D
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R273C/S240R double mutant bound to DNA and MQ: R273C/S240R-DNA-MQ | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Diskin-Posner, Y, Degtjarik, O, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
7B4N
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human wild-type p53DBD bound to DNA and MQ: wt-DNA-MQ (II) | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Diskin-Posner, Y, Degtjarik, O, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
7B49
 
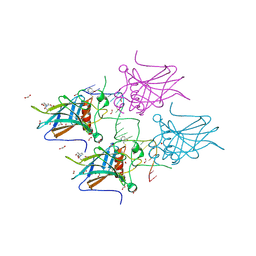 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R273H mutant bound to DNA and MQ: R273H-DNA-MQ | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, 1,2-ETHANEDIOL, ... | | Authors: | Rozenberg, H, Degtjarik, O, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
7B4F
 
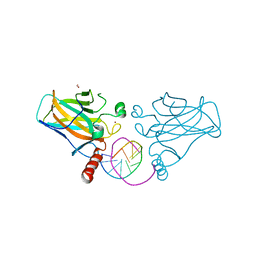 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R282W mutant bound to DNA: R282W-MQ (I) | | Descriptor: | Cellular tumor antigen p53, DNA target, FORMIC ACID, ... | | Authors: | Rozenberg, H, Degtjarik, O, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
2VUK
 
 | |
1OKJ
 
 | |
2WGX
 
 | |
1GIA
 
 | |
4P90
 
 | | Crystal structure of the kinase domain of human PAK1 in complex with compound 15 | | Descriptor: | Serine/threonine-protein kinase PAK 1, [2-chloro-5-(hydroxymethyl)phenyl]{5-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-1H-pyrrolo[2,3-b]pyridin-3-yl}methanone | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-04-01 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Identification and optimisation of 7-azaindole PAK1 inhibitors with improved potency and kinase selectivity.
MEDCHEMCOMM, 2014
|
|
2QZ2
 
 | | Crystal structure of a glycoside hydrolase family 11 xylanase from Aspergillus niger in complex with xylopentaose | | Descriptor: | Endo-1,4-beta-xylanase I, SODIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Vandermarliere, E, Rombouts, S, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
2X0U
 
 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO A 2-amino substituted benzothiazole scaffold | | Descriptor: | 6,7-DIHYDRO[1,4]DIOXINO[2,3-F][1,3]BENZOTHIAZOL-2-AMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Kaar, J.L, Basse, N, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
5K72
 
 | | IRAK4 in complex with Compound 21 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}4,~{N}4-dimethyl-~{N}1-[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]cyclohexane-1,4-diamine | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-25 | | Release date: | 2017-12-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
5K7G
 
 | | IRAK4 in complex with AZ3862 | | Descriptor: | (3~{a}~{S},7~{a}~{R})-1-methyl-5-[4-[[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]amino]cyclohexyl]-3,3~{a},4,6,7,7~{a}-hexahydropyrrolo[3,2-c]pyridin-2-one, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
5JR2
 
 | | Crystal structure of the EphA4 LBD in complex with APYd3 peptide inhibitor | | Descriptor: | APYd3 peptide, Ephrin type-A receptor 4, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Olson, E.J, Pasquale, E.B, Dawson, P.E, Riedl, S.J. | | Deposit date: | 2016-05-05 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modifications of a Nanomolar Cyclic Peptide Antagonist for the EphA4 Receptor To Achieve High Plasma Stability.
Acs Med.Chem.Lett., 7, 2016
|
|
5K76
 
 | | IRAK4 in complex with Compound 28 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-(4-morpholin-4-ylcyclohexyl)-5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Ferguson, A.D. | | Deposit date: | 2016-05-25 | | Release date: | 2017-12-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
