7DUN
 
 | |
2YFN
 
 | | galactosidase domain of alpha-galactosidase-sucrose kinase, AgaSK | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GALACTOSIDASE-SUCROSE KINASE AGASK, GLYCEROL, ... | | Authors: | Sulzenbacher, G, Bruel, L, Tison-Cervera, M, Pujol, A, Nicoletti, C, Perrier, J, Galinier, A, Ropartz, D, Fons, M, Pompeo, F, Giardina, T. | | Deposit date: | 2011-04-07 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Agask, a Bifunctional Enzyme from the Human Microbiome Coupling Galactosidase and Kinase Activities
J.Biol.Chem., 286, 2011
|
|
7L6F
 
 | | The empty AAV11 capsid | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7DTU
 
 | | Human Calcium-Sensing Receptor bound with L-Trp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Extracellular calcium-sensing receptor, ... | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Liu, L, Sun, D.M, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
2YQ6
 
 | | Structure of Bcl-xL bound to BimSAHB | | Descriptor: | BCL-2-LIKE PROTEIN 1, BIM BETA 5, GLYCEROL | | Authors: | Smith, B.J, Czabotar, P.E. | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Stabilizing the Pro-Apoptotic Bimbh3 Helix (Bimsahb) Does not Necessarily Enhance Affinity or Biological Activity.
Acs Chem.Biol., 8, 2013
|
|
7E36
 
 | | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkanesulfonate monooxygenase SsuD/methylene tetrahydromethanopterin reductase-like flavin-dependent oxidoreductase (Luciferase family), ... | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis.
Nat Commun, 12, 2021
|
|
2YGH
 
 | | SAM-I riboswitch with a G2nA mutation in the Kink turn in complex with S-adenosylmethionine | | Descriptor: | POTASSIUM ION, S-ADENOSYLMETHIONINE, SAM-I RIBOSWITCH, ... | | Authors: | Schroeder, K, Daldrop, P, Lilley, D. | | Deposit date: | 2011-04-17 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RNA Tertiary Interactions in a Riboswitch Stabilize the Structure of a Kink Turn.
Structure, 19, 2011
|
|
5MUE
 
 | | Self-assembled alpha-Tocopherol Transfer Protein Nanoparticles Promote Vitamin E Delivery Across an Endothelial Barrier | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, Alpha-tocopherol transfer protein, CHLORIDE ION, ... | | Authors: | Stocker, A, Aeschimann, W. | | Deposit date: | 2017-01-13 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Self-assembled alpha-Tocopherol Transfer Protein Nanoparticles Promote Vitamin E Delivery Across an Endothelial Barrier.
Sci Rep, 7, 2017
|
|
7DTT
 
 | | Human Calcium-Sensing Receptor bound with calcium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Sun, D.M, Liu, L, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
2OKN
 
 | | Crystal Strcture of Human Prolidase | | Descriptor: | HYDROGENPHOSPHATE ION, MANGANESE (II) ION, Xaa-Pro dipeptidase | | Authors: | Mueller, U, Niesen, F.H, Roske, Y, Goetz, F, Behlke, J, Buessow, K, Heinemann, U, Protein Structure Factory (PSF) | | Deposit date: | 2007-01-17 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Human Prolidase: The Molecular Basis of PD Disease.
To be Published
|
|
2YPD
 
 | | Crystal structure of the Jumonji domain of human Jumonji domain containing 1C protein | | Descriptor: | 2-(ETHYLMERCURI-THIO)-BENZOIC ACID, PROBABLE JMJC DOMAIN-CONTAINING HISTONE DEMETHYLATION PROT EIN 2C, SODIUM ION | | Authors: | Vollmar, M, Johansson, C, Krojer, T, Berridge, G, Burgess-Brown, N, Strain-Damerell, C, Froese, S, Williams, E, Goubin, S, Coutandin, D, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Jumonji Domain of Human Jumonji Domain Containing 1C Protein
To be Published
|
|
3HDS
 
 | | Crystal structure of 4-methylmuconolactone methylisomerase in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-methylmuconolactone methylisomerase, short peptide ASWSA | | Authors: | Marin, M, Heinz, D.W, Pieper, D.H, Klink, B.U. | | Deposit date: | 2009-05-07 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure and catalytic mechanism of 4-methylmuconolactone methylisomerase
J.Biol.Chem., 284, 2009
|
|
2YQ1
 
 | |
7E35
 
 | | Crystal structure of the SARS-CoV-2 papain-like protease (PLPro) C112S mutant bound to compound S43 | | Descriptor: | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Non-structural protein 3, ZINC ION | | Authors: | Liu, J, Wang, Y, Xu, X, Pan, L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2.
Cell Chem Biol, 28, 2021
|
|
2JRZ
 
 | | Solution structure of the Bright/ARID domain from the human JARID1C protein. | | Descriptor: | Histone demethylase JARID1C | | Authors: | Koehler, C, Bishop, S, Dowler, E.F, Diehl, A, Schmieder, P, Leidert, M, Sundstrom, M, Arrowsmith, C.H, Wiegelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Backbone and sidechain 1H, 13C and 15N resonance assignments of the Bright/ARID domain from the human JARID1C (SMCX) protein.
Biomol.Nmr Assign., 2, 2008
|
|
3HEK
 
 | |
3HEI
 
 | | Ligand Recognition by A-Class Eph Receptors: Crystal Structures of the EphA2 Ligand-Binding Domain and the EphA2/ephrin-A1 Complex | | Descriptor: | Ephrin type-A receptor 2, Ephrin-A1 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-05-08 | | Release date: | 2009-06-30 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
7E9V
 
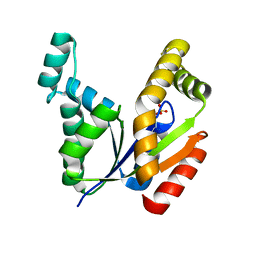 | | The Crystal Structure of human UMP-CMP kinase from Biortus. | | Descriptor: | SULFATE ION, UMP-CMP kinase | | Authors: | Wang, F, Lin, D, Wang, R, Wei, X, Shen, Z, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of human UMP-CMP kinase from Biortus.
To Be Published
|
|
5MVY
 
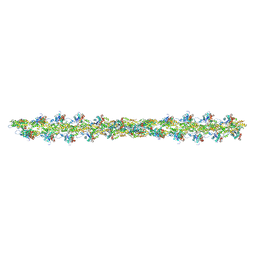 | | Thin Filament at low calcium concentration | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Paul, D.M, Squire, J.M, Morris, E.P. | | Deposit date: | 2017-01-17 | | Release date: | 2018-02-14 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (28.4 Å) | | Cite: | Relaxed and active thin filament structures; a new structural basis for the regulatory mechanism.
J. Struct. Biol., 197, 2017
|
|
3HG1
 
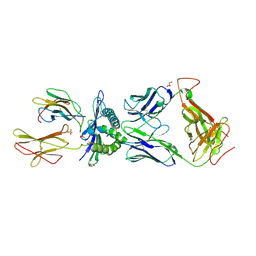 | | Germline-governed recognition of a cancer epitope by an immunodominant human T cell receptor | | Descriptor: | Beta-2-microglobulin, CANCER/MART-1, GLYCEROL, ... | | Authors: | Cole, D.K, Yuan, F, Rizkallah, P.J, Miles, J.J, Gostick, E, Price, D.A, Gao, G.F, Jakobsen, B.K, Sewell, A.K. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-28 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor.
J.Biol.Chem., 284, 2009
|
|
5MWU
 
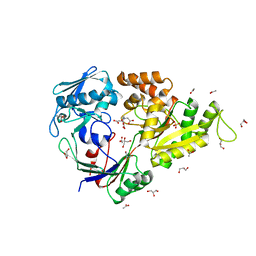 | | Crystal structure of the periplasmic nickel-binding protein NikA from Escherichia coli in complex with Ru(bpza)(CO)2Cl | | Descriptor: | ACETATE ION, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Cavazza, C, Lopez, S, Rondot, L, Iannello, M, Boeri-Erba, E, Burzlaff, N, Strinitz, F, Jorge-Robin, A, Marchi-Delapierre, C, Menage, S. | | Deposit date: | 2017-01-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efficient conversion of alkenes to chlorohydrins by a Ru-based artificial enzyme
To Be Published
|
|
2YHE
 
 | | Structure determination of the stereoselective inverting sec- alkylsulfatase Pisa1 from Pseudomonas sp. | | Descriptor: | SEC-ALKYL SULFATASE, SULFATE ION, ZINC ION | | Authors: | Kepplinger, B, Faber, K, Macheroux, P, Schober, M, Knaus, T, Wagner, U.G. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Mechanism of an Inverting Alkylsulfatase from Pseudomonas Sp. Dsm6611 Specific for Secondary Alkylsulfates.
FEBS J., 279, 2012
|
|
7E7B
 
 | | Cryo-EM structure of the SARS-CoV-2 furin site mutant S-Trimer from a subunit vaccine candidate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | Authors: | Zheng, S, Ma, J. | | Deposit date: | 2021-02-25 | | Release date: | 2021-03-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of S-Trimer, a subunit vaccine candidate for COVID-19.
J.Virol., 95, 2021
|
|
3HF8
 
 | | Crystal structure of human tryoptophan hydroxylase type 1 with bound LP-533401 and Fe | | Descriptor: | 4-{2-amino-6-[(1R)-2,2,2-trifluoro-1-(3'-fluorobiphenyl-4-yl)ethoxy]pyrimidin-4-yl}-L-phenylalanine, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Tari, L.W, Swanson, R.V, Hunter, M.J. | | Deposit date: | 2009-05-11 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Inhibition of Novel Tryptophan Hydroxylase Inhibitors Revealed by Co-crystal Structures and Kinetic Analysis.
Curr Chem Genomics, 4, 2010
|
|
2OAE
 
 | | Crystal structure of rat dipeptidyl peptidase (DPPIV) with thiazole-based peptide mimetic #31 | | Descriptor: | Dipeptidyl peptidase 4, N-{[(3S,5S)-5-(1,3-THIAZOLIDIN-3-YLCARBONYL)PYRROLIDIN-3-YL]METHYL}-1,3-THIAZOLE-4-CARBOXAMIDE, SULFATE ION | | Authors: | Longenecker, K.L, Shuai, Q, Patel, J, Wiedeman, P. | | Deposit date: | 2006-12-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pyrrolidine-constrained phenethylamines: The design of potent, selective, and pharmacologically efficacious dipeptidyl peptidase IV (DPP4) inhibitors from a lead-like screening hit.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
