2HAH
 
 | | The structure of FIV 12S protease in complex with TL-3 | | Descriptor: | Protease, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Heaslet, H, Lin, Y.C, Elder, J.H, Stout, C.D. | | Deposit date: | 2006-06-12 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an FIV/HIV chimeric protease complexed with the broad-based inhibitor, TL-3.
Retrovirology, 4, 2007
|
|
1E7S
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase K140R | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Zuccotti, S, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
7G0E
 
 | | Crystal Structure of human FABP5 in complex with 2-[(3-ethoxycarbonyl-4,5,6,7-tetrahydro-1-benzothiophen-2-yl)carbamoyl]cyclopentene-1-carboxylic acid, i.e. SMILES C1CCC2=C(C1)C(=C(S2)NC(=O)C1=C(C(=O)O)CCC1)C(=O)OCC with IC50=1.1 microM | | Descriptor: | 2-{[3-(ethoxycarbonyl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5P91
 
 | | humanized rat catechol O-methyltransferase in complex with 5-(4-fluorophenyl)-2,3-dihydroxy-N-[2-[5-(2-methylpyridin-4-yl)-4H-1,2,4-triazol-3-yl]ethyl]benzamide at 1.20A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-(4-fluorophenyl)-2,3-dihydroxy-N-[2-[5-(2-methylpyridin-4-yl)-4H-1,2,4-triazol-3-yl]ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of a COMT complex
To be published
|
|
3ET1
 
 | |
1PS3
 
 | | Golgi alpha-mannosidase II in complex with kifunensine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase II, ... | | Authors: | Shah, N, Kuntz, D.A, Rose, D.R. | | Deposit date: | 2003-06-20 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of Kifunensine and 1-Deoxymannojirimycin Binding to Class I and II alpha-Mannosidases Demonstrates Different Saccharide Distortions in Inverting and Retaining Catalytic Mechanisms
Biochemistry, 42, 2003
|
|
3ZXK
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIAXHD3, alpha-L-arabinofuranose-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OSG
 
 | | The structure of protozoan parasite Trichomonas vaginalis Myb2 in complex with MRE-1-12 DNA | | Descriptor: | 5'-D(*AP*AP*AP*TP*AP*TP*CP*GP*TP*TP*AP*T)-3', 5'-D(*AP*TP*AP*AP*CP*GP*AP*TP*AP*TP*TP*T)-3', MYB21 | | Authors: | Jiang, I, Tsai, C.K, Chen, S.C, Wang, S.H, Amiraslanov, I, Chang, C.F, Wu, W.J, Tai, J.H, Liaw, Y.C, Huang, T.H. | | Deposit date: | 2010-09-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Molecular basis of the recognition of the ap65-1 gene transcription promoter elements by a Myb protein from the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 39, 2011
|
|
3TWV
 
 | | Crystal structure of ARC4 from human Tankyrase 2 in complex with peptide from human NUMA1 (chimeric peptide) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, ... | | Authors: | Guettler, S, Sicheri, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease.
Cell(Cambridge,Mass.), 147, 2011
|
|
1LMB
 
 | | REFINED 1.8 ANGSTROM CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Beamer, L.J, Pabo, C.O. | | Deposit date: | 1991-11-05 | | Release date: | 1991-11-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A crystal structure of the lambda repressor-operator complex.
J.Mol.Biol., 227, 1992
|
|
4NYQ
 
 | | In-vivo crystallisation (midguts of a viviparous cockroach) and structure at 1.2 A resolution of a glycosylated, lipid-binding, lipocalin-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Coussens, N.P, Gallat, F.-X, Ramaswamy, S, Yagi, K, Tobe, S.S, Stay, B, Chavas, L.M.G. | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a heterogeneous, glycosylated, lipid-bound, in vivo-grown protein crystal at atomic resolution from the viviparous cockroach Diploptera punctata.
Iucrj, 3, 2016
|
|
1E4E
 
 | | D-alanyl-D-lacate ligase | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roper, D.I. | | Deposit date: | 2000-07-03 | | Release date: | 2001-06-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular basis of vancomycin resistance in clinically relevant Enterococci: crystal structure of D-alanyl-D-lactate ligase (VanA).
Proc. Natl. Acad. Sci. U.S.A., 97, 2000
|
|
7G1N
 
 | | Crystal Structure of human FABP4 in complex with rac-(1R,2R)-2-[[3-(3-methyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl]cyclohexane-1-carboxylic acid, i.e. SMILES C1(=C(C2=C(S1)CCCC2)C1=NC(=NO1)C)NC(=O)[C@@H]1[C@H](CCCC1)C(=O)O with IC50=0.365 microM | | Descriptor: | (1R,2R)-2-{[(3M)-3-(3-methyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}cyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Neidhart, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3FPU
 
 | |
2H06
 
 | |
2AHS
 
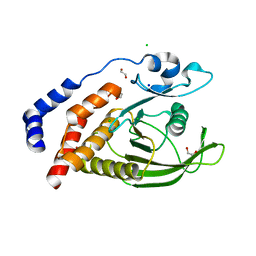 | | Crystal Structure of the Catalytic Domain of Human Tyrosine Receptor Phosphatase Beta | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Receptor-type tyrosine-protein phosphatase beta, ... | | Authors: | Ugochukwu, E, Eswaran, J, Barr, A, Gileadi, O, Sobott, F, Burgess, N, Ball, L, Bray, J, von Delft, F, Debreczeni, J, Bunkoczi, G, Turnbull, A, Das, S, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
1DPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 11), INSULIN B CHAIN (PH 11), ... | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
7KI0
 
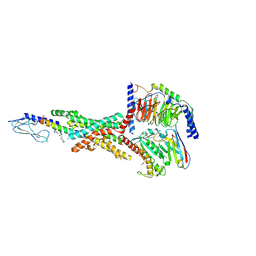 | | Semaglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs protein | | Descriptor: | 17-amino-10-oxo-3,6,12,15-tetraoxa-9-azaheptadecan-1-oic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Belousoff, M.J, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2020-10-22 | | Release date: | 2021-08-04 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure and dynamics of semaglutide- and taspoglutide-bound GLP-1R-Gs complexes.
Cell Rep, 36, 2021
|
|
2APC
 
 | |
1ELP
 
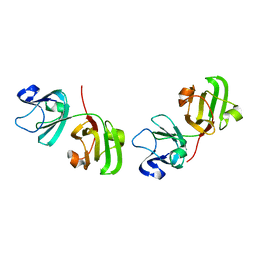 | | GAMMA-D CRYSTALLIN STRUCTURE AT 1.95 A RESOLUTION | | Descriptor: | GAMMA-D CRYSTALLIN | | Authors: | Chirgadze, Yu.N, Driessen, H.P.C, Wright, G, Slingsby, C, Hay, R.E, Lindley, P.F. | | Deposit date: | 1995-12-20 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of bovine eye lens gammaD (gammaIIIb)-crystallin at 1.95 A.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
3IKF
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 717, imidazo[2,,1-b][1,3]thiazol-6-ylmethanol | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
4RZ7
 
 | | Crystal Structure of PVX_084705 with bound PCI32765 | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, UNKNOWN ATOM OR ION, cGMP-dependent protein kinase, ... | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal Structure of PVX_084705 with bound PCI32765
To be Published
|
|
4JJA
 
 | |
5OXG
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with LDN-212854 | | Descriptor: | 1,2-ETHANEDIOL, 5-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1, ... | | Authors: | Williams, E.P, Sorrell, F.J, Kopec, J, Nowak, R.P, Kupinska, K, von Delft, F, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for the potent and selective binding of LDN-212854 to the BMP receptor kinase ALK2.
Bone, 109, 2018
|
|
3I7U
 
 | | Crystal structure of AP4A hydrolase (aq_158) from Aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AP4A hydrolase, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Nakagawa, N, Sekar, K, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Free and ATP-bound structures of Ap(4)A hydrolase from Aquifex aeolicus V5
Acta Crystallogr.,Sect.D, 66, 2010
|
|
