4F2Q
 
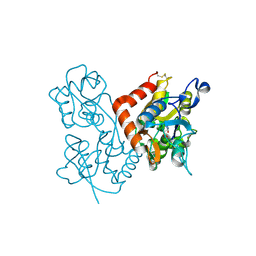 | | Quisqualate bound to the D655A mutant of the ligand binding domain of GluA3 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | The loss of an electrostatic contact unique to AMPA receptor ligand binding domain 2 slows channel activation.
Biochemistry, 51, 2012
|
|
4F3B
 
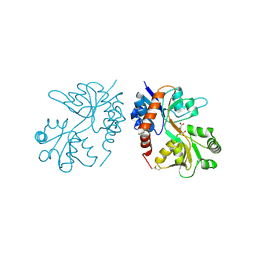 | |
4F31
 
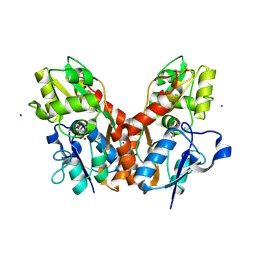 | |
4F39
 
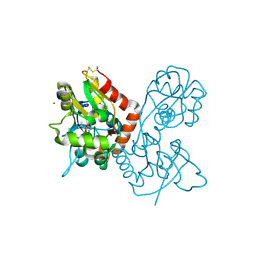 | | Kainate bound to the ligand binding domain of GluA3 | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | The loss of an electrostatic contact unique to AMPA receptor ligand binding domain 2 slows channel activation.
Biochemistry, 51, 2012
|
|
4F22
 
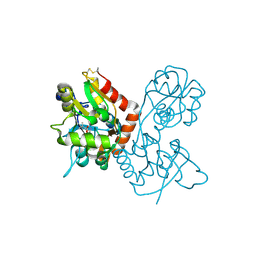 | |
8BN2
 
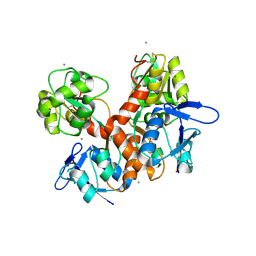 | | Crystal structure of the ligand-binding domain (LBD) of human iGluR Delta-1 (GluD1) in complex with D-Serine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Heroven, C, Malinauskas, T, Aricescu, A.R. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | GluD1 binds GABA and controls inhibitory plasticity.
Science, 382, 2023
|
|
8BN5
 
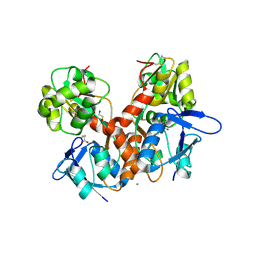 | | Crystal structure of the ligand-binding domain (LBD) of human iGluR Delta-1 (GluD1) in complex with GABA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Heroven, C, Malinauskas, T, Aricescu, A.R. | | Deposit date: | 2022-11-12 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | GluD1 binds GABA and controls inhibitory plasticity.
Science, 382, 2023
|
|
8BLJ
 
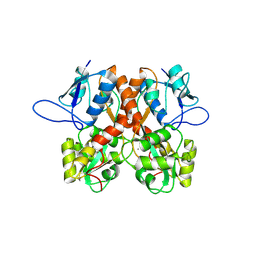 | | Crystal structure of the ligand-binding domain (LBD) of human iGluR Delta-1 (GluD1), apo state | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Heroven, C, Malinauskas, T, Aricescu, A.R. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | GluD1 binds GABA and controls inhibitory plasticity.
Science, 382, 2023
|
|
8BSU
 
 | | Crystal structure of the kainate receptor GluK3-H523A ligand binding domain in complex with kainate and the positive allosteric modulator BPAM344 at 2.9A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2022-11-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Small-molecule positive allosteric modulation of homomeric kainate receptors GluK1-3: development of screening assays and insight into GluK3 structure.
Febs J., 291, 2024
|
|
8BST
 
 | | Crystal structure of the kainate receptor GluK3-H523A ligand binding domain in complex with kainate at 2.7A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2022-11-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule positive allosteric modulation of homomeric kainate receptors GluK1-3: development of screening assays and insight into GluK3 structure.
Febs J., 291, 2024
|
|
8P3V
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 3 | | Descriptor: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3U
 
 | |
8P3T
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 1 | | Descriptor: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | Authors: | Zhang, D, Krieger, J, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3W
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 4 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3X
 
 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Q
 
 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
4G8N
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist G8M | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]cyclobutanecarboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Kastrup, J.S, Frydenvang, K, Gajhede, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological and structural characterization of conformationally restricted (S)-glutamate analogues at ionotropic glutamate receptors.
J.Struct.Biol., 180, 2012
|
|
8P3Z
 
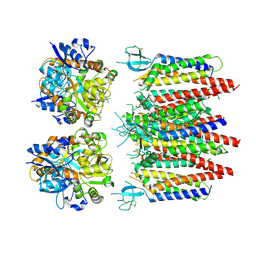 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 2 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Y
 
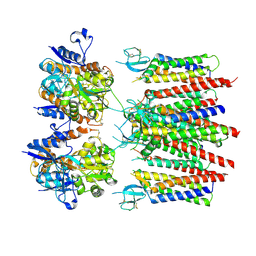 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 3 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3S
 
 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
4G8M
 
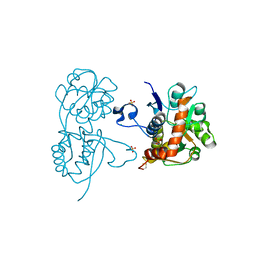 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the agonist CBG-IV at 2.05A resolution | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]cyclobutanecarboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Juknaite, L, Frydenvang, K, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pharmacological and structural characterization of conformationally restricted (S)-glutamate analogues at ionotropic glutamate receptors.
J.Struct.Biol., 180, 2012
|
|
8C2H
 
 | | Transmembrane domain of active state homomeric GluA1 AMPA receptor in tandem with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8F0O
 
 | |
8C1P
 
 | | Active state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CYCLOTHIAZIDE, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C2I
 
 | | Transmembrane domain of resting state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
