9B3P
 
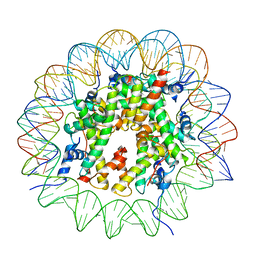 | |
4HRH
 
 | | Crystal Structure of p11-Annexin A2(N-terminal) Fusion Protein in Complex with SMARCA3 Peptide | | Descriptor: | Helicase-like transcription factor, Protein S100-A10, Annexin A2, ... | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2012-10-27 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | SMARCA3, a Chromatin-Remodeling Factor, Is Required for p11-Dependent Antidepressant Action.
Cell(Cambridge,Mass.), 152, 2013
|
|
8BOT
 
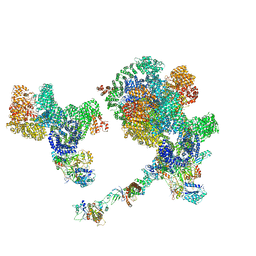 | |
7NFC
 
 | | Cryo-EM structure of NHEJ super-complex (dimer) | | Descriptor: | DNA (27-MER), DNA (28-MER), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
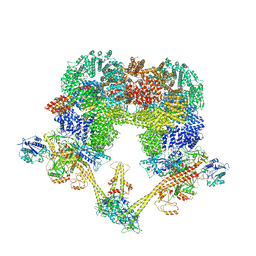
8QFC
 
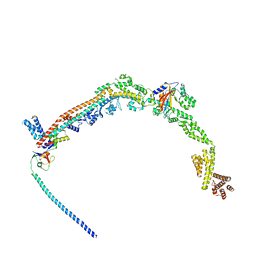 | | UFL1 E3 ligase bound 60S ribosome | | Descriptor: | 60S ribosomal protein L10a, CDK5 regulatory subunit-associated protein 3, DDRGK domain-containing protein 1, ... | | Authors: | Makhlouf, L, Zeqiraj, E, Kulathu, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
8P81
 
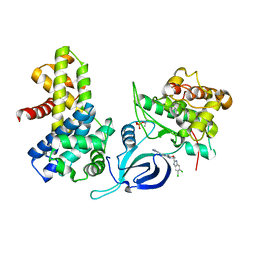 | | Crystal structure of human Cdk12/Cyclin K in complex with inhibitor SR-4835 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, ~{N}-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methyl]-9-(1-methylpyrazol-4-yl)-2-morpholin-4-yl-purin-6-amine | | Authors: | Anand, K, Schmitz, M, Geyer, M. | | Deposit date: | 2023-05-31 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The reversible inhibitor SR-4835 binds Cdk12/cyclin K in a noncanonical G-loop conformation.
J.Biol.Chem., 300, 2023
|
|
8P0D
 
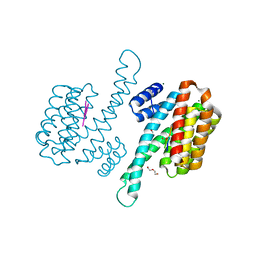 | | Human 14-3-3 sigma in complex with human MDM2 peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Roversi, P, Ward, J, Doveston, R, Kwon, H, Romartinez Alonso, B. | | Deposit date: | 2023-05-10 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Characterizing the protein-protein interaction between MDM2 and 14-3-3 sigma ; proof of concept for small molecule stabilization.
J.Biol.Chem., 300, 2024
|
|
8FVI
 
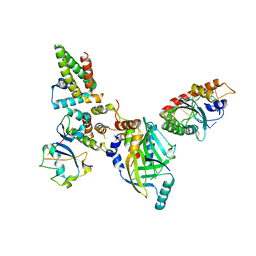 | | Human APOBEC3H bound to HIV-1 Vif in complex with CBF-beta, ELOB, ELOC, and CUL5 | | Descriptor: | Core-binding factor subunit beta, Cullin 5, DNA dC->dU-editing enzyme APOBEC-3H, ... | | Authors: | Ito, F, Alvarez-Cabrera, A.L, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H.
Nat Commun, 14, 2023
|
|
6Z6P
 
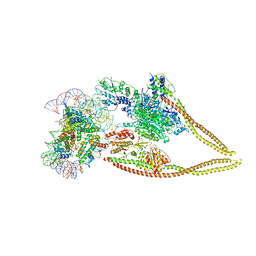 | | HDAC-PC-Nuc | | Descriptor: | DNA (145-MER), HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
8B9B
 
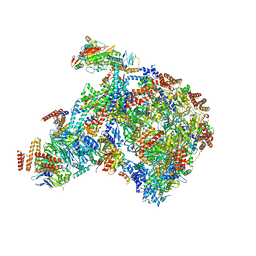 | |
8B9C
 
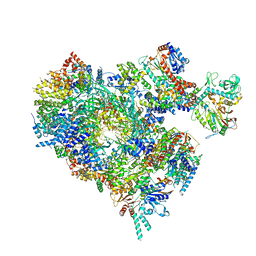 | | S. cerevisiae pol alpha - replisome complex | | Descriptor: | Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA polymerase alpha catalytic subunit A, ... | | Authors: | Jones, M.L, Yeeles, J.T.P. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | How Pol alpha-primase is targeted to replisomes to prime eukaryotic DNA replication.
Mol.Cell, 83, 2023
|
|
8B9A
 
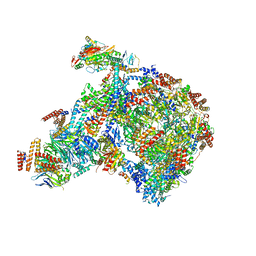 | |
3MWV
 
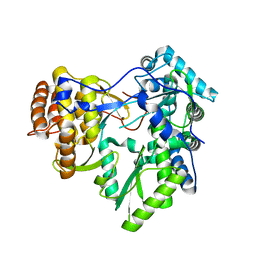 | | Crystal structure of HCV NS5B polymerase | | Descriptor: | Genome polyprotein | | Authors: | Coulombe, R. | | Deposit date: | 2010-05-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of ligand bioactive conformation in the discovery of potent indole-diamide inhibitors of the hepatitis C virus NS5B.
J.Am.Chem.Soc., 132, 2010
|
|
4HRG
 
 | |
8AVG
 
 | | Cryo-EM structure of mouse Elp123 with bound SAM | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
1H2T
 
 | | Structure of the human nuclear cap-binding-complex (CBC) in complex with a cap analogue m7GpppG | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, 80 KDA NUCLEAR CAP BINDING PROTEIN, ... | | Authors: | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2002-08-16 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
1H2V
 
 | | Structure of the human nuclear cap-binding-complex (CBC) | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 80 KDA NUCLEAR CAP BINDING PROTEIN | | Authors: | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2002-08-16 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
1N54
 
 | | Cap Binding Complex m7GpppG free | | Descriptor: | 20 kDa nuclear cap binding protein, 80 kDa nuclear cap binding protein, GLYCEROL | | Authors: | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | Deposit date: | 2002-11-04 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
1AWI
 
 | |
1H2U
 
 | | Structure of the human nuclear cap-binding-complex (CBC) in complex with a cap analogue m7GpppG | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, 80 KDA NUCLEAR CAP BINDING PROTEIN, ... | | Authors: | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2002-08-16 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
6Z1P
 
 | |
1A2B
 
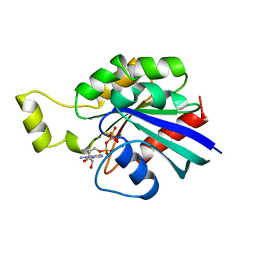 | | HUMAN RHOA COMPLEXED WITH GTP ANALOGUE | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, TRANSFORMING PROTEIN RHOA | | Authors: | Ihara, K, Muraguchi, S, Kato, M, Shimizu, T, Shirakawa, M, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1997-12-26 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue.
J.Biol.Chem., 273, 1998
|
|
7PMK
 
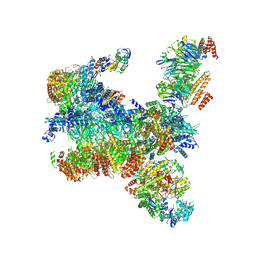 | | S. cerevisiae replisome-SCF(Dia2) complex bound to double-stranded DNA (conformation I) | | Descriptor: | Cell division control protein 45,Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA helicase, ... | | Authors: | Jenkyn-Bedford, M, Yeeles, J.T.P, Deegan, T.D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
7NFE
 
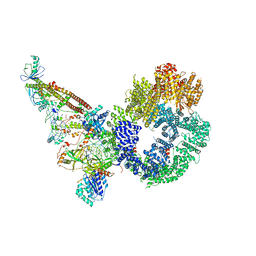 | | Cryo-EM structure of NHEJ super-complex (monomer) | | Descriptor: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*CP*TP*AP*TP*TP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(P*TP*AP*AP*TP*AP*AP*TP*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*AP*G)-3'), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-06 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
7NYF
 
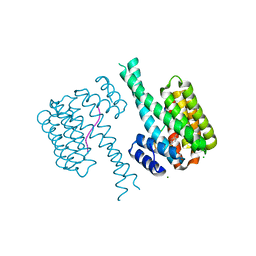 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-131 | | Descriptor: | 14-3-3 protein sigma, 4-[(3~{S})-3-oxidanylpiperidin-1-yl]sulfonylbenzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-22 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
