7E9K
 
 | | Crystal Structure of POMGNT2 in complex with UDP and mono-mannosyl peptide (379Man long peptide) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2021-03-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan.
Genes Cells, 26, 2021
|
|
3MO0
 
 | | Human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E11 | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Chang, Y, Horton, J.R, Cheng, X. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
3MOD
 
 | | Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced IgG) with 11 aa gp41 MPER-derived peptide | | Descriptor: | ANTI-HIV-1 ANTIBODY 2F5 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 2F5 LIGHT CHAIN, gp41 MPER-derived peptide | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
5QJ6
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1614545742 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, MAGNESIUM ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJJ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z24758179 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJY
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z94597856 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1Y6G
 
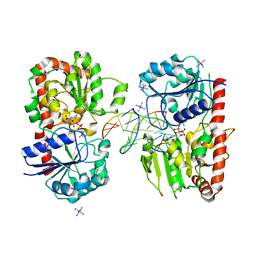 | | alpha-glucosyltransferase in complex with UDP and a 13_mer DNA containing a HMU base at 2.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*T)-3', 5'-D(*GP*AP*TP*AP*CP*TP*(5HU)P*AP*GP*AP*TP*AP*G)-3', ... | | Authors: | Lariviere, L, Sommer, N, Morera, S. | | Deposit date: | 2004-12-06 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase.
J.Mol.Biol., 352, 2005
|
|
4FEW
 
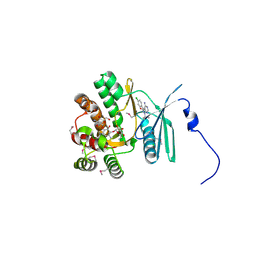 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor pyrazolopyrimidine PP2 | | Descriptor: | 1-TERT-BUTYL-3-(4-CHLORO-PHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
3MOE
 
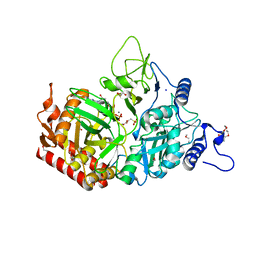 | |
5QOT
 
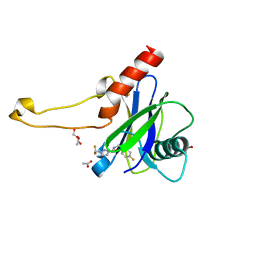 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1592710382 | | Descriptor: | 1,2-ETHANEDIOL, 1-(difluoromethyl)-N-[(4-fluorophenyl)methyl]-1H-pyrazole-3-carboxamide, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QP8
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with PB1787571279 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1YHY
 
 | | Crystal structure of Arabidopsis thaliana Acetohydroxyacid synthase In Complex With A Sulfonylurea Herbicide, Metsulfuron methyl | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ETHYL DIHYDROGEN DIPHOSPHATE, ... | | Authors: | McCourt, J.A, Pang, S.S, King-Scott, J, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1Y2A
 
 | | Structure of mammalian importin bound to the non-classical PLSCR1-NLS | | Descriptor: | Importin alpha-2 Subunit, decamer fragment of Phospholipid scramblase 1 | | Authors: | Chen, M.-H, Ben-Efraim, I, Mitrousis, G, Walker-Kopp, N, Sims, P.J, Cingolani, G. | | Deposit date: | 2004-11-22 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phospholipid Scramblase 1 Contains a Nonclassical Nuclear Localization Signal with Unique Binding Site in Importin alpha
J.Biol.Chem., 280, 2005
|
|
1QWY
 
 | | Latent LytM at 1.3 A resolution | | Descriptor: | ZINC ION, peptidoglycan hydrolase | | Authors: | Odintsov, S.G, Sabala, I, Marcyjaniak, M, Bochtler, M. | | Deposit date: | 2003-09-03 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Latent LytM at 1.3A resolution.
J.Mol.Biol., 335, 2004
|
|
7G5F
 
 | | Crystal Structure of rat Autotaxin in complex with 1H-benzotriazol-5-yl-[rac-(1R,2R,6S,7S)-9-[4-(cyclopropylmethoxy)naphthalene-2-carbonyl]-4,9-diazatricyclo[5.3.0.02,6]decan-4-yl]methanone, i.e. SMILES N1(C[C@H]2[C@@H](C1)[C@H]1[C@@H]2CN(C1)C(=O)c1ccc2c(c1)N=NN2)C(=O)c1cc(c2c(c1)cccc2)OCC1CC1 with IC50=0.00985223 microM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
2OPB
 
 | | Structure of K57A hPNMT with inhibitor 3-fluoromethyl-7-thiomorpholinosulfonamide-THIQ and AdoHcy | | Descriptor: | (3R)-3-(FLUOROMETHYL)-7-(THIOMORPHOLIN-4-YLSULFONYL)-1,2,3,4-TETRAHYDROISOQUINOLINE, PHOSPHATE ION, Phenylethanolamine N-methyltransferase, ... | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2007-01-28 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase.
J.Med.Chem., 50, 2007
|
|
4INS
 
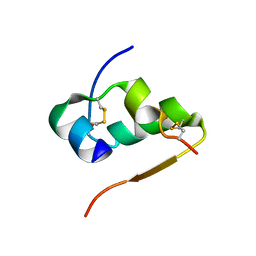 | | THE STRUCTURE OF 2ZN PIG INSULIN CRYSTALS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | INSULIN (CHAIN A), INSULIN (CHAIN B), ZINC ION | | Authors: | Dodson, G.G, Dodson, E.J, Hodgkin, D.C, Isaacs, N.W, Vijayan, M. | | Deposit date: | 1989-07-10 | | Release date: | 1990-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of 2Zn pig insulin crystals at 1.5 A resolution.
Philos.Trans.R.Soc.London,Ser.B, 319, 1988
|
|
3CR5
 
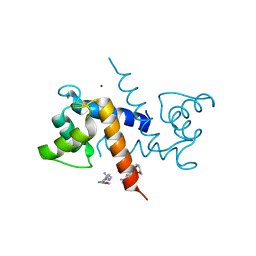 | | X-ray structure of bovine Pnt-Zn(2+),Ca(2+)-S100B | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, CALCIUM ION, Protein S100-B, ... | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
3M7W
 
 | | Crystal Structure of Type I 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 in Covalent Complex with Dehydroquinate | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.
J.Biol.Chem., 286, 2011
|
|
4OP8
 
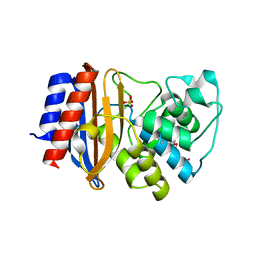 | | Room temperature crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying G238S mutation | | Descriptor: | CALCIUM ION, SULFATE ION, TEM-94 ES-beta-lactamase, ... | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
4OPQ
 
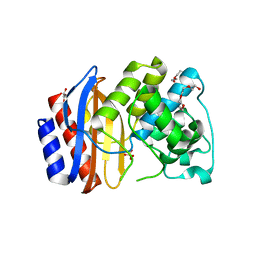 | | Room temperature crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying R164S/G238S mutations | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-06 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
5QP9
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z100435060 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQR
 
 | | PanDDA analysis group deposition -- Crystal Structure of human ALAS2A in complex with Z1171217421 | | Descriptor: | 1-[(2-methyl-1,3-thiazol-5-yl)methyl]piperazine, 5-aminolevulinate synthase, erythroid-specific, ... | | Authors: | Bezerra, G.A, Foster, W, Bailey, H, Shrestha, L, Krojer, T, Talon, R, Brandao-Neto, J, Douangamath, A, Nicola, B.B, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Brennan, P.E, Yue, W.W. | | Deposit date: | 2019-05-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QJU
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z906021418 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-2-(propan-2-yl)pyrimidine-4-carboxamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4IOS
 
 | | Structure of phage TP901-1 RBP (ORF49) in complex with nanobody 11. | | Descriptor: | BPP, GLYCEROL, Llama nanobody 11 | | Authors: | Desmyter, A, Farenc, C, Mahony, J, Spinelli, S, Bebeacua, C, Blangy, S, Veesler, D, van Sinderen, D, Cambillau, C. | | Deposit date: | 2013-01-08 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
