2KXX
 
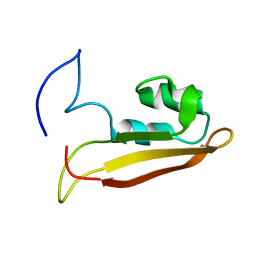 | | NMR Structure of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex | | Descriptor: | Small protein A | | Authors: | Kim, K, Okon, M, Escobar, E, Kang, H, McIntosh, L, Paetzel, M. | | Deposit date: | 2010-05-13 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex.
Biochemistry, 50, 2011
|
|
2Q1Z
 
 | |
4RUL
 
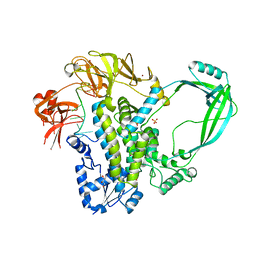 | | Crystal structure of full-length E.Coli topoisomerase I in complex with ssDNA | | Descriptor: | DNA topoisomerase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Chen, B, Tse-Dinh, Y.C. | | Deposit date: | 2014-11-20 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I.
Nucleic Acids Res., 43, 2015
|
|
4TT3
 
 | | The Pathway of Binding of the Intrinsically Disordered Mitochondrial Inhibitor Protein to F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2014-06-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4AST
 
 | |
4TSF
 
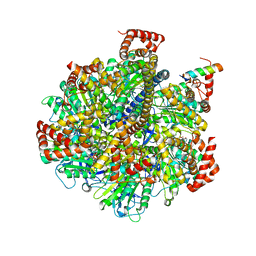 | | The Pathway of Binding of the Intrinsically Disordered Mitochondrial Inhibitor Protein to F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1ML5
 
 | | Structure of the E. coli ribosomal termination complex with release factor 2 | | Descriptor: | 30S 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Klaholz, B.P, Pape, T, Zavialov, A.V, Myasnikov, A.G, Orlova, E.V, Vestergaard, B, Ehrenberg, M, van Heel, M. | | Deposit date: | 2002-08-30 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structure of the Escherichia coli ribosomal termination complex with release factor 2
Nature, 421, 2003
|
|
1EN5
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Y34F MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1U60
 
 | | MCSG APC5046 Probable glutaminase ybaS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable glutaminase ybaS | | Authors: | Chang, C, Cuff, M.E, Joachimiak, A, Savchenko, A, Edwards, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
2AKQ
 
 | | The structure of bovine B-lactoglobulin A in crystals grown at very low ionic strength | | Descriptor: | Beta-lactoglobulin variant A | | Authors: | Adams, J.J, Anderson, B.F, Norris, G.E, Creamer, L.K, Jameson, G.B. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bovine beta-lactoglobulin (variant A) at very low ionic strength
J.Struct.Biol., 154, 2006
|
|
1HQF
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH N-HYDROXY-L-ARGININE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, N-OMEGA-HYDROXY-L-ARGININE | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
2NBT
 
 | | NEURONAL BUNGAROTOXIN, NMR, 10 STRUCTURES | | Descriptor: | NEURONAL BUNGAROTOXIN | | Authors: | Oswald, R.E, Sutcliffe, M.J, Bamberger, M, Loring, R.H, Braswell, E, Dobson, C.M. | | Deposit date: | 1997-10-29 | | Release date: | 1998-03-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuronal bungarotoxin determined by two-dimensional NMR spectroscopy: calculation of tertiary structure using systematic homologous model building, dynamical simulated annealing, and restrained molecular dynamics.
Biochemistry, 31, 1992
|
|
2LEU
 
 | | HIGH RESOLUTION 1H NMR STUDY OF LEUCOCIN A IN 90% AQUEOUS TRIFLUOROETHANOL (TFE) (0.1% TFA), 18 STRUCTURES | | Descriptor: | LEUCOCIN A | | Authors: | Gallagher, N.L.F, Sailer, M, Niemczura, W.P, Nakashima, T.T, Stiles, M.E, Vederas, J.C. | | Deposit date: | 1997-05-20 | | Release date: | 1997-11-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria.
Biochemistry, 36, 1997
|
|
4ZCS
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase in complex with CDP-choline | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Choline-phosphate cytidylyltransferase, [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM | | Authors: | Guca, E, Hoh, F, Guichou, J.-F, Cerdan, R. | | Deposit date: | 2015-04-16 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural determinants of the catalytic mechanism of Plasmodium CCT, a key enzyme of malaria lipid biosynthesis.
Sci Rep, 8, 2018
|
|
1MUE
 
 | | Thrombin-Hirugen-L405,426 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(1-OXIDO-2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUOROPHENYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Nantermet, P.G, Selnick, H.G, Isaacs, R.C, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Stranieri, M.T, Cook, J.J, McMasters, D.R, Pellicore, J.M, Pal, S, Wallace, A.A, Clayton, F.C, Bohn, D, Welsh, D.C, Lynch, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pharmacokinetic optimization of 3-amino-6-chloropyrazinone acetamide thrombin inhibitors. Implementation of P3 pyridine N-oxides to deliver an orally bioavailable series containing P1 N-benzylamides.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
3LI0
 
 | | Crystal structure of the mutant R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
4GBE
 
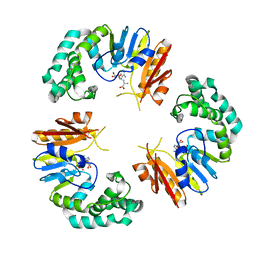 | |
2RGW
 
 | |
6I5M
 
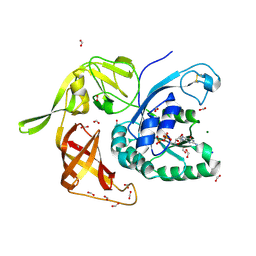 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus in complex with GDP and formate ion | | Descriptor: | FORMIC ACID, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kravchenko, O, Nikonov, O, Gabdulkhakov, A, Stolboushkina, E, Arkhipova, V, Garber, M, Nikonov, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The third structural switch in the archaeal translation initiation factor 2 (aIF2) molecule and its possible role in the initiation of GTP hydrolysis and the removal of aIF2 from the ribosome.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2BS0
 
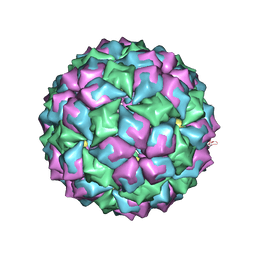 | | MS2 (N87AE89K mutant) - Variant Qbeta RNA hairpin complex | | Descriptor: | 5'-R(*AP*UP*GP*CP*AP*UP*GP*UP*CP*UP *AP*AP*GP*AP*CP*UP*GP*CP*AP*U)-3', COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2RB9
 
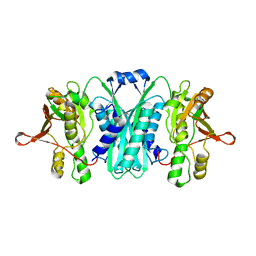 | | Crystal structure of E.coli HypE | | Descriptor: | HypE protein | | Authors: | Asinas, A.E, Rangarajan, E.S, Min, T, Matte, A, Proteau, A, Munger, C, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-09-18 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of [NiFe] hydrogenase maturation protein HypE from Escherichia coli and its interaction with HypF.
J.Bacteriol., 190, 2008
|
|
1P9W
 
 | | Crystal Structure of Vibrio cholerae putative NTPase EpsE | | Descriptor: | General secretion pathway protein E, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ZINC ION | | Authors: | Robien, M.A, Krumm, B.E, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2003-05-12 | | Release date: | 2003-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae
J.Mol.Biol., 333, 2003
|
|
1ELJ
 
 | | THE CRYSTAL STRUCTURE OF LIGANDED MALTODEXTRIN-BINDING PROTEIN FROM PYROCOCCUS FURIOSUS | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Evdokimov, A.G, Anderson, E.D, Routzahn, K.M, Waugh, D.S. | | Deposit date: | 2000-03-13 | | Release date: | 2001-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for oligosaccharide recognition by Pyrococcus furiosus maltodextrin-binding protein.
J.Mol.Biol., 305, 2001
|
|
1EVF
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CYS167 MUTANT OF ESCHERICHIA COLI | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
5I2X
 
 | | Crystal Structure of TPP1 K170del | | Descriptor: | Adrenocortical dysplasia protein homolog | | Authors: | Nandakumar, J, Smith, E. | | Deposit date: | 2016-02-09 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional consequences of a disease mutation in the telomere protein TPP1.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
